I spent the last 24 hours on flights across the international date line with a barrage of emails asking me for my opinion on this recent Dhuli paper.
I was half attentive to it as I didn’t have great connectivity and was frankly more concerned about getting a Barry Young welcoming party at the New Zealand border patrol.
Yes..
To add insult to injury, we had planned a whole family trip down to New Zealand to revisit my old stomping ground. As part of my Pre-Med course at Emory University, I did a semester abroad at University of Otago, Dunedin in 1994 and have been dying to get back to the country. When I heard Jacinda the Jab Jigalo had left office, I figured there might be a ‘shot’ at a safe return.
That was before I got tangled up hosting
‘obfuscated’ New Zealand data and having the TWO (NZMH) illegally take down my Mega.nz drive.Mega.nz has since restored our data but it was not clear to me if the country would try to extend its Barry Young tyranny on other people in the story. So I spent the flight preparing my family members for the remote possibility that their father might be whisked away for some questioning upon arrival and armed them with a few attorneys phone numbers in case “ignorance became strength”.
I did put a few of my thoughts on the paper on Twitter.
Knowing I have a bias, I try to look at every paper that supports what I suspect to be happening from the eyes of my worst critics.
In this case Elizabeth Bik has probably read my thread and mirrored much of my concerns in her PubPeer bashing while she sprinkled in her hatred of people in the supplement industry. Recall, Elizabeths company was raided by the FBI for selling fecal microbiome sequencing snake oil. Now she props herself up as some defender of good science with hit pieces on any entity in the COViD space that sells a vitamin.
As you can see, I’m not convinced this paper moves the needle on the integration topic and I predicted this would arm the Jab Jigalos with a victorious slam dunk after many people in our field circulated it as a potential jab kill shot. I do appreciate the authors looking at this topic. Most run from it.
So lets look at how this Alley-oop plays out.
Elizabeth raises interesting points about the authors conflicts of interest and the fact that the authors have close relationships with the Journal and to some supplement companies.
That’s very interesting.
This has never bothered Elizabeth before!
These same issues exist on the Corman-Drosten PCR paper where the authors had undisclosed interests in PCR testing companies (later acquired by Roche for millions). The authors were also the editors of the journal and somehow managed a 24 hour peer review on a PCR paper that had no internal controls.
When ever this double standard is brought to Elizabeth Biks attention, she goes silent. In fact, I think she mobilized a mob of people on PubPeer to attack our critique of these conflicts of interest. So she really doesn’t care about these conflicts. She just needs to weaponize them against the papers from people outside of her tribe.
Its a form of Supplement Derangement Syndrome.
Its very clear to many scientists in the field that Elizabeth isn’t an honest broker. She is a hitman (hitwoman or whatever her pronoun clan calls her). She is likely hired to attack papers that challenge the pharma narrative.
Everyone took note when Elizabeth emerged unscathed from the FBI raid of the Fecal Theronos of her time. The company she worked for (ubiome) was raided by the FBI for selling snake oil fecal sequencing and many suspect she is now a hired hand for the establishment. Its rare for such a crash and burn to be heralded as beacon of scientific myth-busters without the establishment encouraging her rebirth.
The Dhuli papers can be seen here-
https://www.europeanreview.org/wp/wp-content/uploads/013-019-2.pdf
The supplement has the PCR and sequencing data.
https://www.europeanreview.org/wp/wp-content/uploads/supplementary-data.pdf
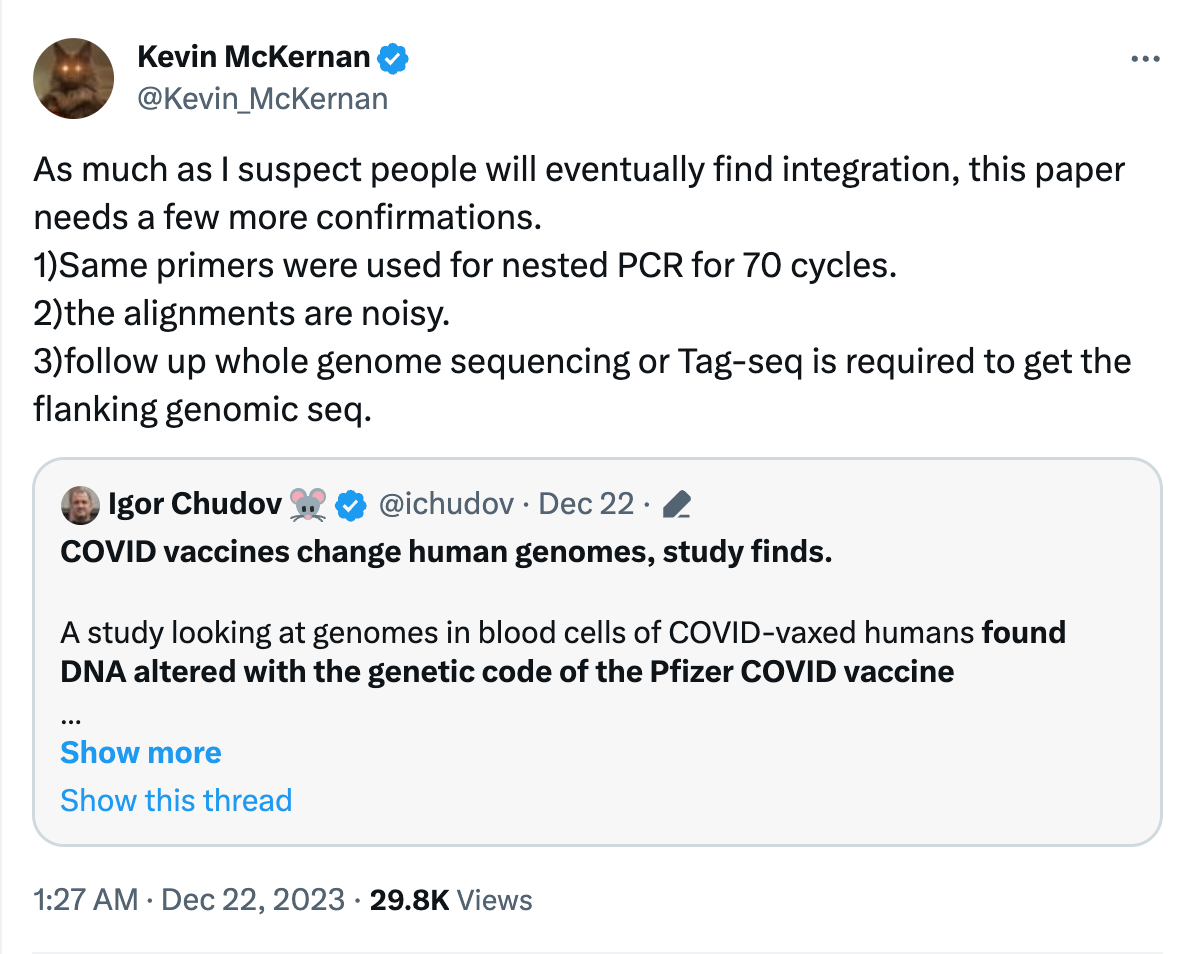
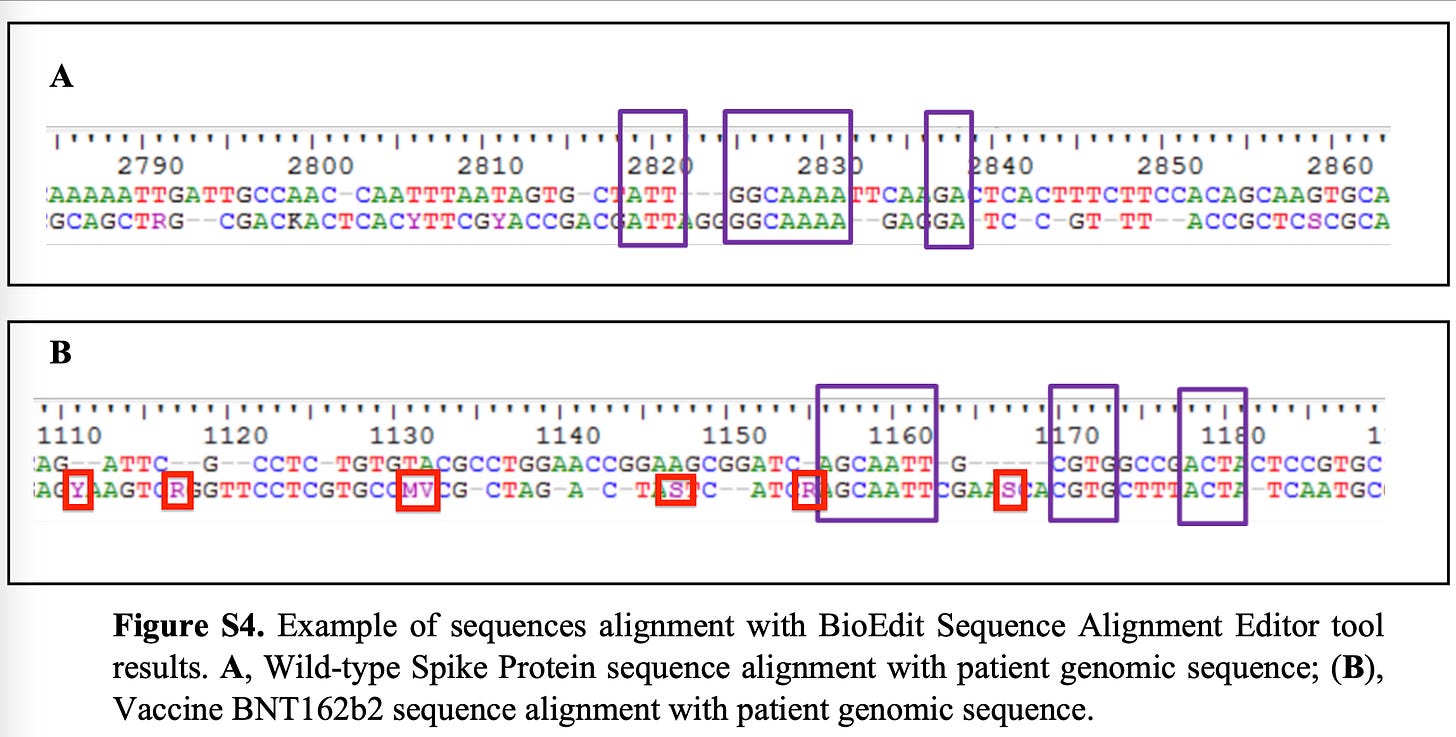
Here is where my criticisms emerge. All of the bases in Red Boxes are highlighted by me. These are not ATCGs. They are IUPAC codes for degenerate bases.
R = Purine or A/G.
Y= Pyrimidine or C/T
When a sequencing read presents you with IUPAC bases it means the sequencer cant discern the actual base and has to make degenerate base call.
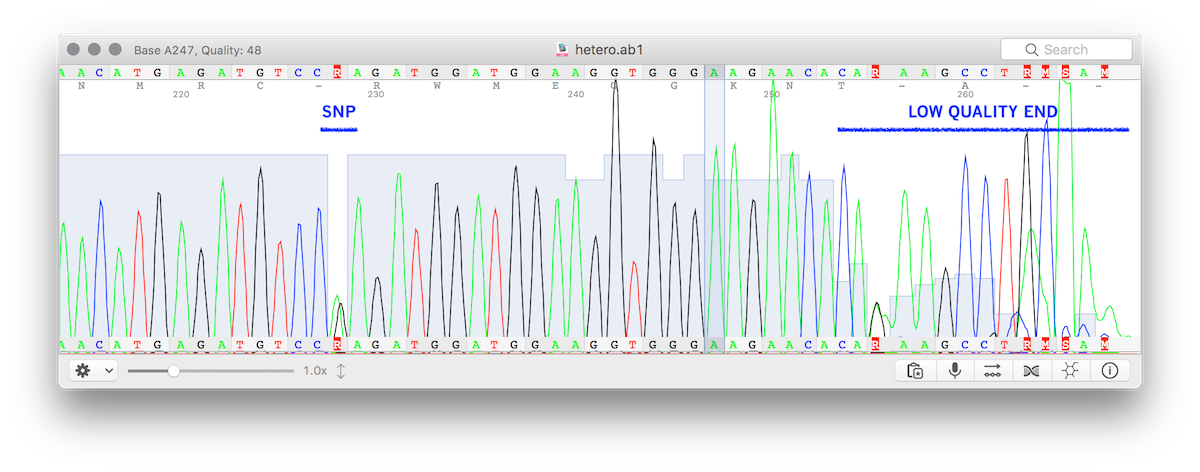
This is often a sign that the Sanger sequencing is a mess. You can see this in the alignments. They are not contiguous alignments but contain only short sequence stretches of homology seen in purple boxes. It would be helpful in these controversial scenarios to put the Sanger traces live for everyone to review. There is other dimensionality to Sanger data that is lost when you distill the whole trace into ATCGs. The quality scores are lost. You can see a high quality ‘R’ as a SNP in the below trace and a bunch of other low quality regions of the reads producing R, M, S calls that are likely noise. The quality scores are light blue scale you see decay towards the right side of the trace.
So without the actual quality scores and traces being public, Im inclined to think this is a PCR artifact that happens to generate some short sequences that align to your hypothesis.
How was the Sanger Sequencing derived?
From a PCR product. They called this Nested PCR but it really was not nested and they did not try to hide this despite Elizabeth trying to school them on their own admission. This is common behavior amongst the Jab jigalos. They take a limitation you admit to in your paper and then weaponize as if its something you tried to hide from the reader.
From Dhuli et al. Note- they spell this out precisely and there is reason to attempt 2 rounds of PCR. Between the 2 PCR protocols, you can purify the amplicons to remove the small off target primer dimers and repeat the PCR in attempt to clean it up. Not ideal but also not at all suspect as the authors were very transparent about this.
What does Elizabeth turn this into…
Her own little captain obvious scolding.
Don’t you love it when Captain Obvious marches in to enlighten you about what you actually did and told her you did! This is typical behavior from low citation ‘debunker’ clowns. They routinely play this trick on the Fluorometry work in our PrePrints. We spell out the cross-talk limitations which I’m quite suspect they would have even recognized without our transparency, and then try to spin it into some attempt of ours to manipulate the reader.
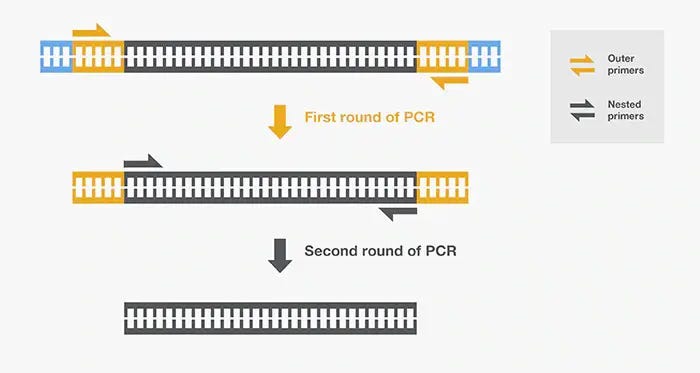
So, what is nested PCR-
Nested PCR uses 2 rounds of PCR with inner and outer primers. The use of 2 different primer sets allows you to double confirm you are amplifying your target. When you perform ‘nested PCR’ with non-nested PCR primers, you are really just asking for PCR to over amplify what was generated with 35 cycles of your 1st PCR.
There is one justifiable reason to do this. If your 1st round of PCR presents some small off target bands, you can gel purify the band you think is the right target and attempt to amplify your isolated band again. The gel purification should remove gDNA or RNA contaminations that may be the source of the off target amplification.
So this is fair game as long as you disclose this is what you did and Dhuli authors were quite clear about this.
How can they improve this data.
Just showing you have a spike like amplicon doesnt tell you it came from a gDNA integration event.
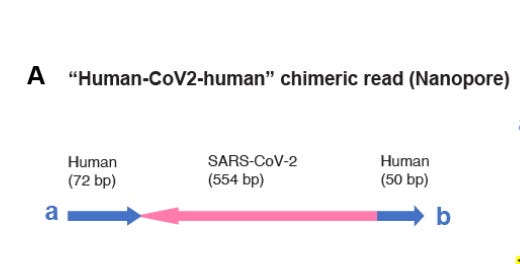
Lets look at what Zhang et al did to confirm integration of SARs-CoV-2. You’ll need to dig into their supplement to see this.
You can see that they capture Human-Spike-Human sequence with Oxford Nanopore. Not just spike, but human-spike-human sequence. One approach is to use Whole genome sequencing. This is an expensive way to confirm this as you need to sequence very deeply across the whole genome to find it and not all cells will be integrated.
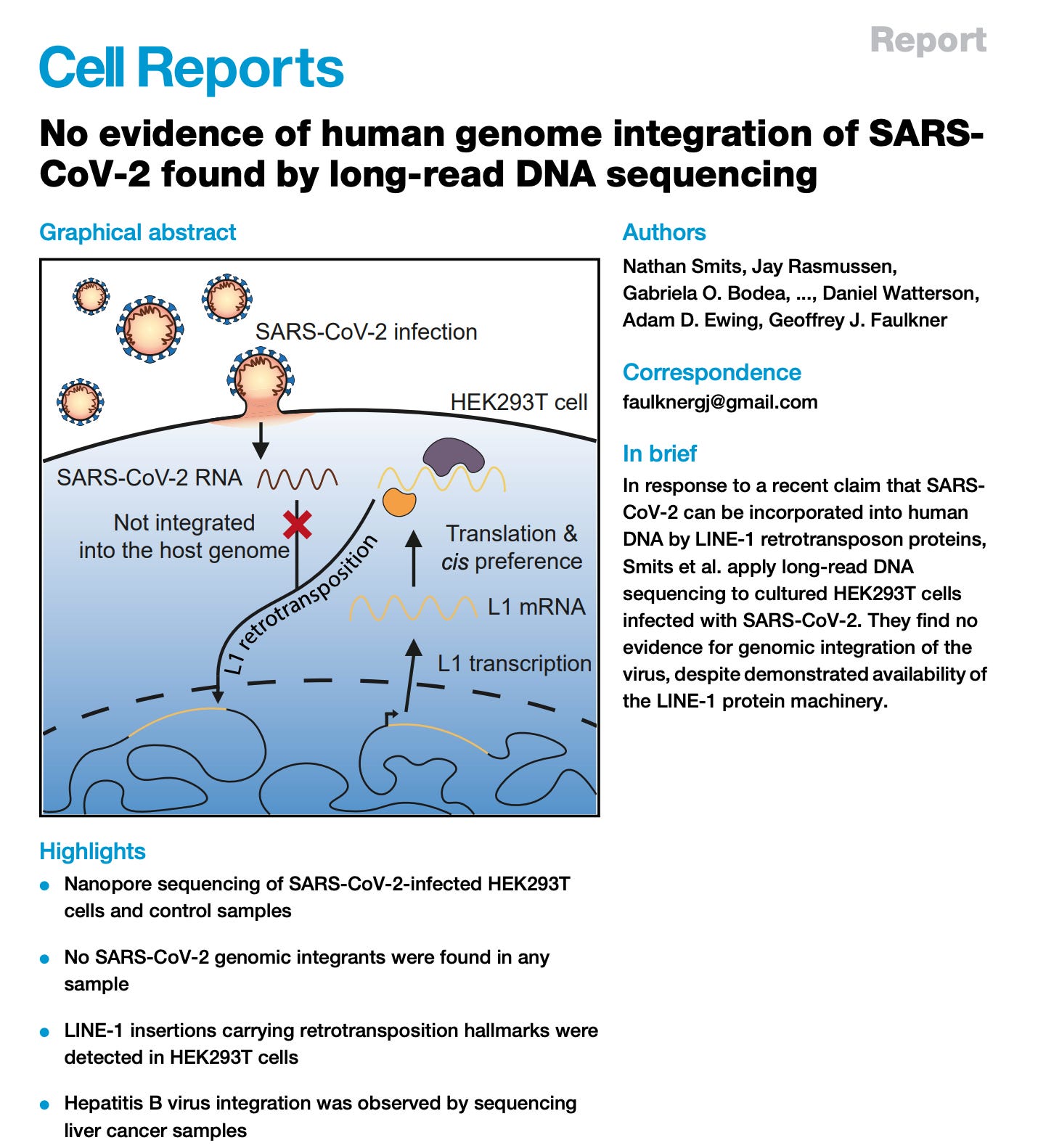
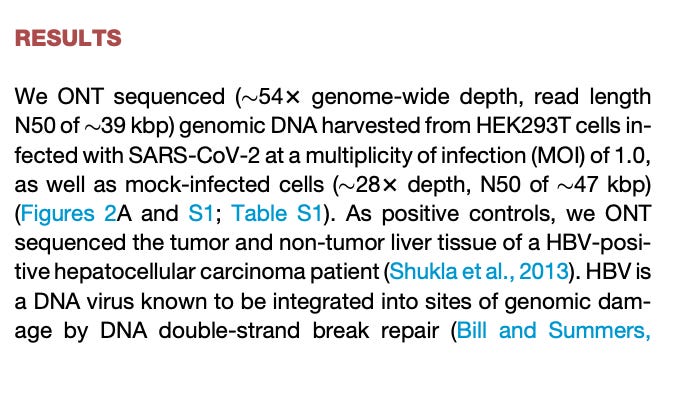
A small minority will be integrated and thus what often happens is people don’t find the samples that have this as they only sequence 28X-54X deep in a given patient. This shallow sequencing has led to a few papers that have tried to unconvincingly refute the Zhang paper. The Zhang et al PNAS results were challenged with Smits et al in Cell.
You can see that they came to this conclusion by not looking very hard. 28X-54X coverage is what is required to detect integration that happens in 50-100% of the cells in the sample. If less than 1% of the cells are integrated, you’ll have less than 1 read detecting the integration event at 50X coverage. The fact that this overt deception made it into Cell while papers raising the alarm are relegated to ‘predatory’ journals like MDPI is a statement in and of itself. I don’t subscribe to ‘predatory’ label of MDPI. This is a comment usually voiced by economic illiterates and we’ll save that topic for another post.
Zhang put an end to this debate with their MDPI paper in 2023. In this paper they more carefully examine their results next to the concerns raised by other scientists. The main complaint was that their results could have been a template switching artifact in the process of amplifying RNA or DNA. There are some other rare artifacts that can occur making sequencing libraries where two distant pieces of DNA get fused in the ligation step used to put your sequencing primers onto the DNA.
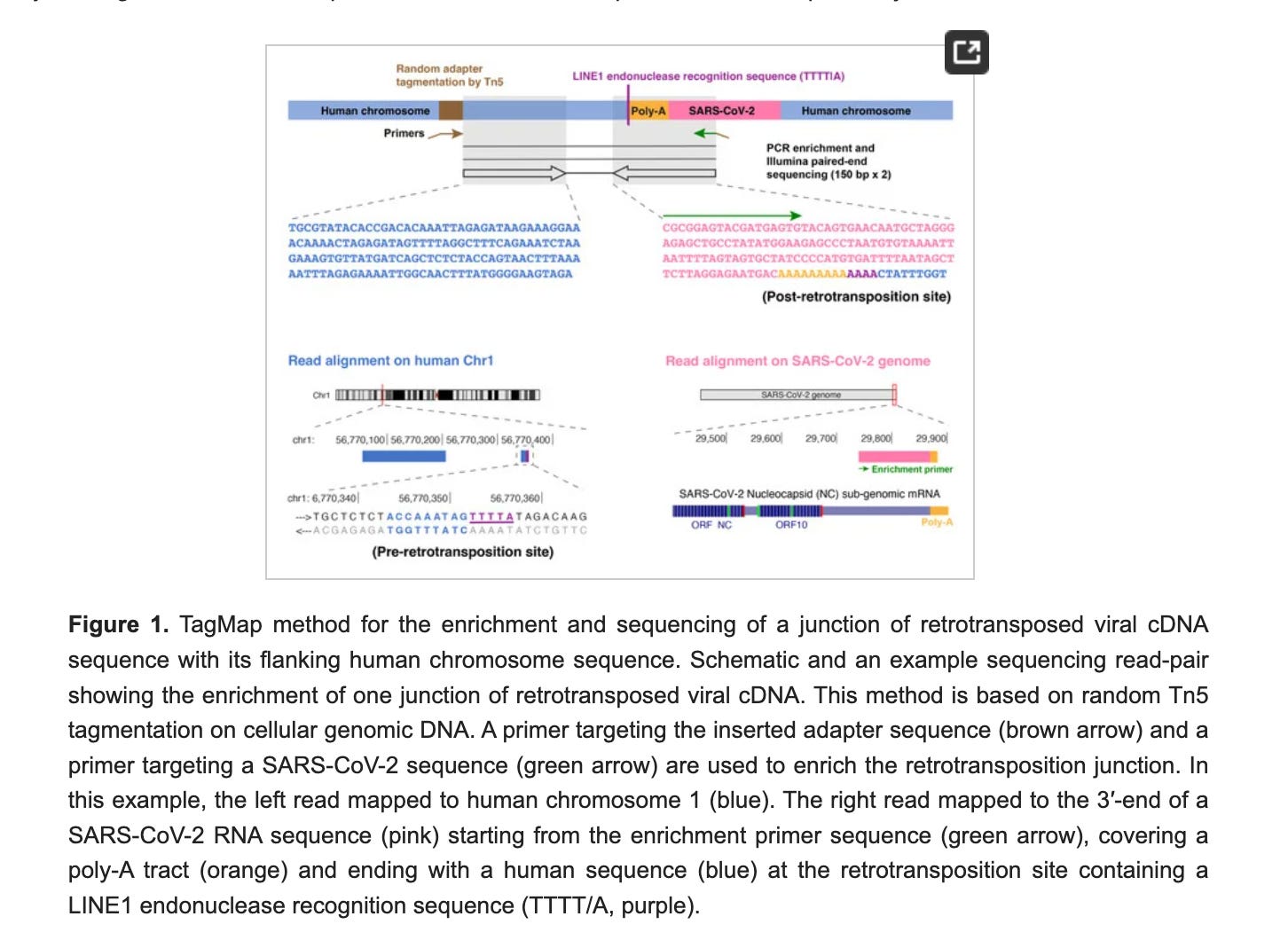
Zhang et al developed a more sensitive method called TagMap that enables you to amplify the regions with integration events. This enables one to find needles in a haystack.
The way they do this is they randomly integrate the genome with Tn5 transposon sequences so that these Tn5 integrations occur once every 500-1000 bases. Once the genome is decorated with a known Tn5 primer site every ~500 bases, they can PCR with a spike primer and a Tn5 primer to enrich the regions with Spike and human junctions from the background. This is a very clever way to pull out low frequency events from the 6 billion bases in each of the ~100,000 cells they survey.
Other papers that have touched on this topic are below but I think the best lay person read on the topic is Rudi’s interview on the topic here.
In my opinion, we have clear evidence that mRNA from SARs-CoV-2 has been shown to integrate and no one to date has adequately addressed the vaccine integration issue. Zhang et al tried to address this with a mock RNA but this RNA didn’t contain modified RNAs or SV40 nuclear localization sequences.
This does shoot a major elephant in the room that Peter Marks, Paul Offit and other Jab jigalo bullies recite… The mRNA cant get to the nucleus. If SARs-CoV-2 mRNA can integrate then there is no reason a modified RNA can’t. And once you know the modRNA is contaminated with DNA that has Nuclear Targeting Sequences, you know the gig is up. The ‘Cant get to the nucleus’ argument has already been annihilated by experts in the field with SARs-CoV-2 but they don’t want to acknowledge this.
This is why Marks, Offit and all of the factchecker cited pimps NEVER address the SV40 NTS. They all stop their argument at this DNA is getting destroyed in the cytosol.
I have no doubt integration will be found if funded. I suspect the funding from the conflicted NIH will only flow to those scientists looking to debunk this $400M vaccine royalty threat.
Even once this is found, the goal posts will once again shift to this integration being found in a tiny number of cells and being clinically meaningless. Just as DNA contamination was originally denied, then normalized. Just as Frameshifting was predicted, denied, then found and normalized.
This reminds me of a great read from another AnandAmigo-
The process is the punishment.
Enjoy the Holiday. I will likely be off line for a trek or two. I have a preprint consuming me at the moment which should emerge in the new year.
https://www.frontiersin.org/articles/10.3389/fmicb.2021.676693/full
https://retrovirology.biomedcentral.com/articles/10.1186/s12977-021-00578-w
https://www.mdpi.com/1999-4915/15/3/629






Happy Christmas and New Year Kevin. Throughout 2023 you, and your team, have done some amazing research that has profound implications for all humanity. ““Integrity is telling myself the truth. And honesty is telling the truth to other people” quote from Spencer Johnson. A very big thank you from me.
Merry Christmas and Happy Hollidays Kevin. It has been a pleasure (and education) following you.