Cannabis Testing and Lawfare in Michigan
A case for transparent public testing data and more frequent secret shopper programs
Our last post focused on grows suing regulators in Oregon. To be fair, a different twist on this story is unfolding in Michigan but the lawsuit is to deny an Aspergillus recall the Michigan Cannabis Regulatory Authority (MI-CRA) attempted to enforce.
Michigan has their largest cannabis testing lab (Viridis) suing the regulators over a $229M recall the MI-CRA declared. In both OR and MI suits, the consumer loses.
A few Oregon grows sued the regulators to remove Aspergillus testing from the state entirely and in the case in Michigan, the lab (Viridis) is suing the regulators to allow ‘allegedly’ Aspergillus contaminated product to get to the market. But only half of this later statement is true as Viridis had 2 testing labs and their Northern lab got lumped in with the recall that should have only targeted the Lansing facility.
This Michigan case is very nuanced as both parties have some valid points, however, this is a case that also could have been avoided with more real time public data transparency.
Given the size of this recall, presumably, if Viridis didn’t step up and fight this, their customers (grows) could sue them for the recall, thus bankrupting the lab. In retrospect this may be viewed as an over reaction from the regulators as it put the lab ‘All-In’ on the issue. It’s best to police the market in bite size increments so businesses can adapt and make change but if a given policing action is to revoke 60% of the product in the state ($229M), lawsuits are going to fly.
There are several prior issues that likely led to this lab going ‘All-In’ against their regulators. For one, the Michigan CRA was known for changing the rules frequently on the labs. These unfunded mandates cost labs considerable money as they had to adapt to new rules, run new validations and in many cases there was no consensus on the scientific justification for the changes. This complaint was listed in the Viridis lawsuit.
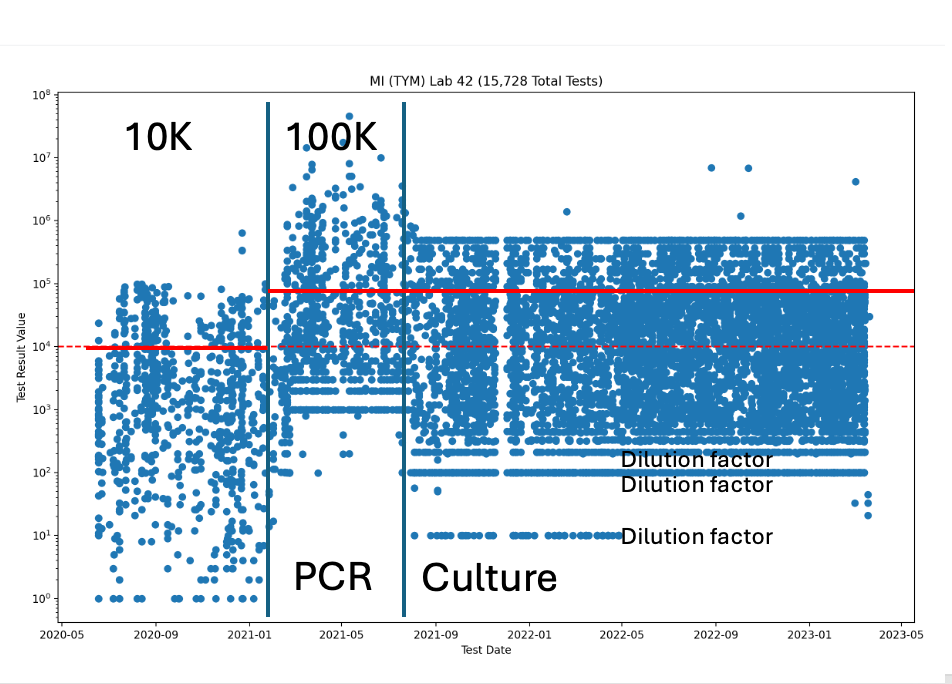
As a reference, just have a look at the Michigan TYM history in Figure 5 below. TYM is first brought in at 10,000 CFU/g action limit, then moved to 100,000 CFU/g in 6 months…. and then an Emergency ERV/AOAC study is required to move everyone to the platform that enjoys a Michigan tax base (Neogen/3M Plating). All of these changes occurred in 18 months and many of our lab partners expressed extreme frustration at this constant change.
The MI-CRA may have exposed themselves legally on all of this change. The Emergency AOAC study was snowballed on the accusation that qPCR was missing TYM counts compared to 3M. 3M’s food testing division is owned by Neogen who is a Lansing based Michigan company. Missing TYM counts meant qPCR ‘could be’ a threat to patients.
When the study completed the data showed that at least one qPCR kit (Medicinal Genomics) was counting more CFU/g than plating! Despite this, plating was declared to be the victor in the study likely because the study was centrally organized by a lab with a Neogen partnership (Steadfast labs). The study was organized with culture based methods assumed to be the gold standard that everything had to be benchmarked to (more on this below).
The MI-CRA should certainly be concerned by the role reversal here (undercounting is bad, except if its from a platform in your tax jurisdiction) but instead they moved ahead with the technology that undercounted (in non-remediated cases).
This is likely playing a role in the Viridis case against the MI-CRA as Viridis is challenging their competency in choosing a gold standard for declaring such a recall. Likewise there is a time delay from when Viridis tested the sample and when the MI-CRA tested it on the shelf. Both labs could be right and microbes simply grew on the shelf. The field needs shelf life testing to have any validity to such lawsuits.
The MI-CRA is claiming no liability and thus no discovery via qualified immunity since Cannabis is federally illegal.
I don’t think its a good idea for regulators to have no skin in the game as they can begin to behave recklessly. Thankfully another judge agreed and dismissed this argument. However, when this case was escalated to federal judges, they dismissed the case on the grounds that cannabis is federally illegal. The case escalated to going directly after the regulators in a personal capacity (whoa) which will set a precedence in the field for very cautious regulators. It’s rare a patient ever sues a regulator but if you over police a grow or a lab quality checking a grow, there is different calculus involved.
It appears the personal lawsuits for Viridis North have now been dismissed but the Lansing case is still underway and a ruling is expected by the end of 2024.
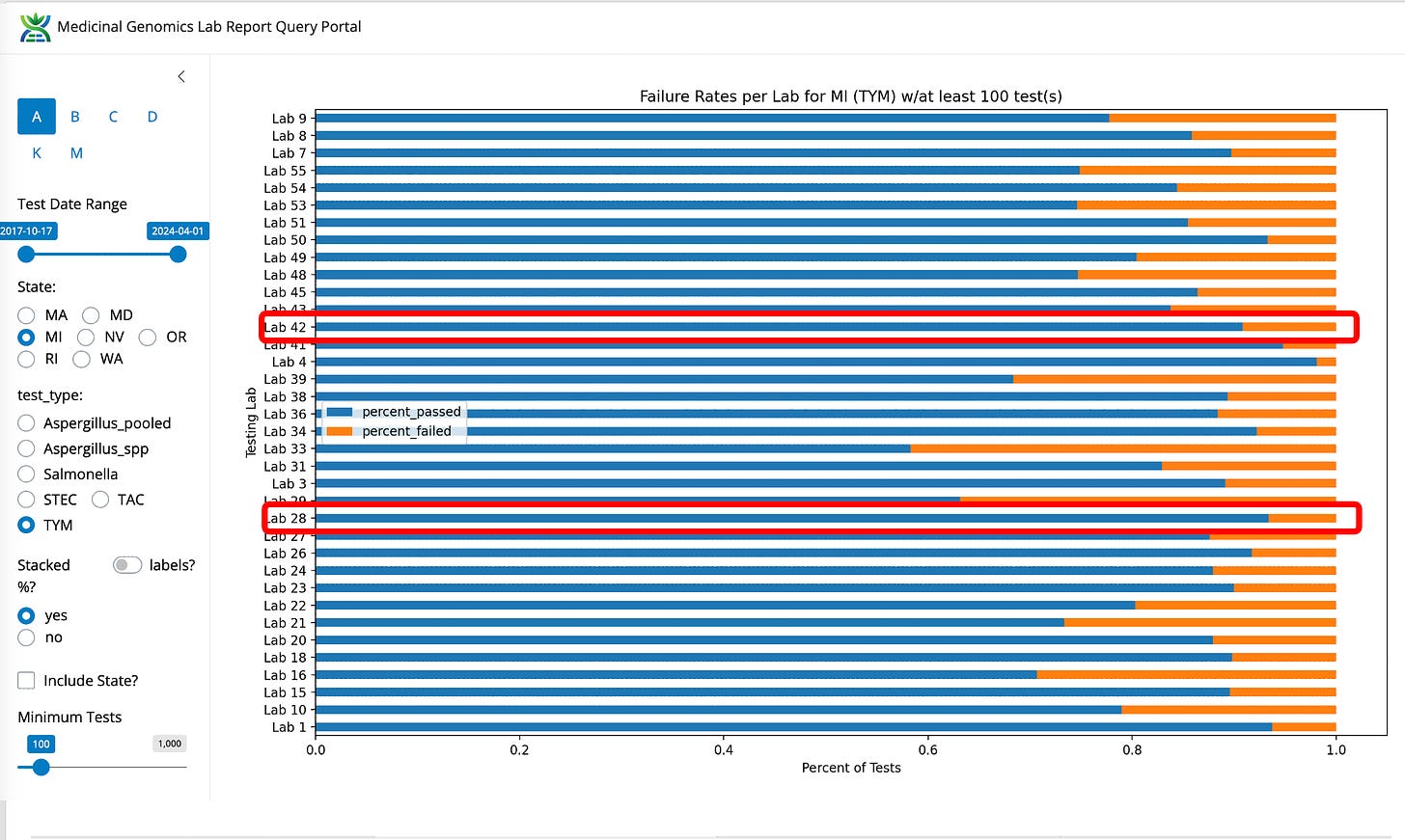
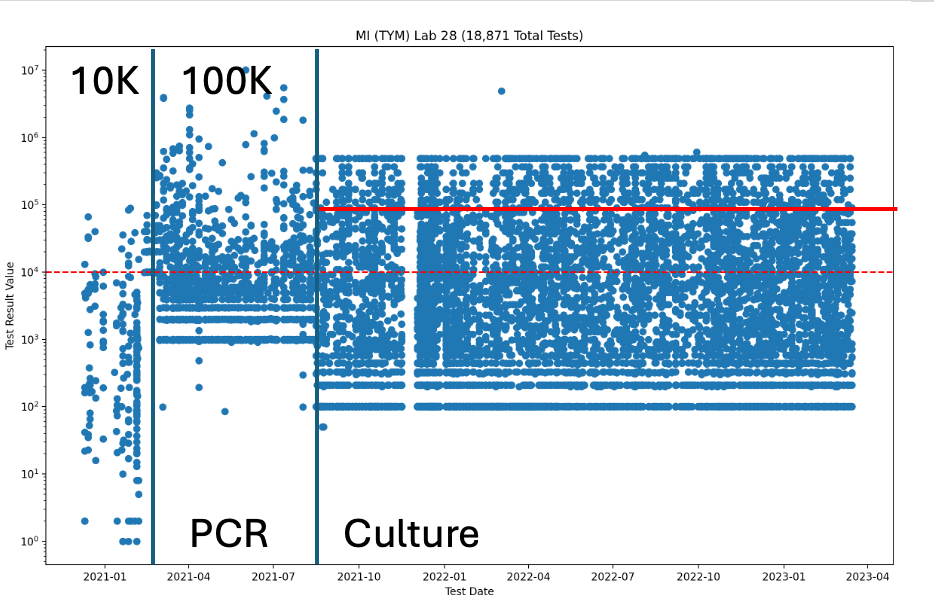
What likely triggered the MI-CRA to take action against Viridis, is that this lab happens to have the highest testing volume in the state and the lowest TYM failure rate. Lab 42 and 28 are noted as LN (Lansing) and BC (Bay City) in the notes section of the Michigan data (graciously provided by Yasha Kahn but the BC and LN assumptions are my own based on their court documents).
Figure 1. TYM fail rates for the State of Michigan normalized by Lab.
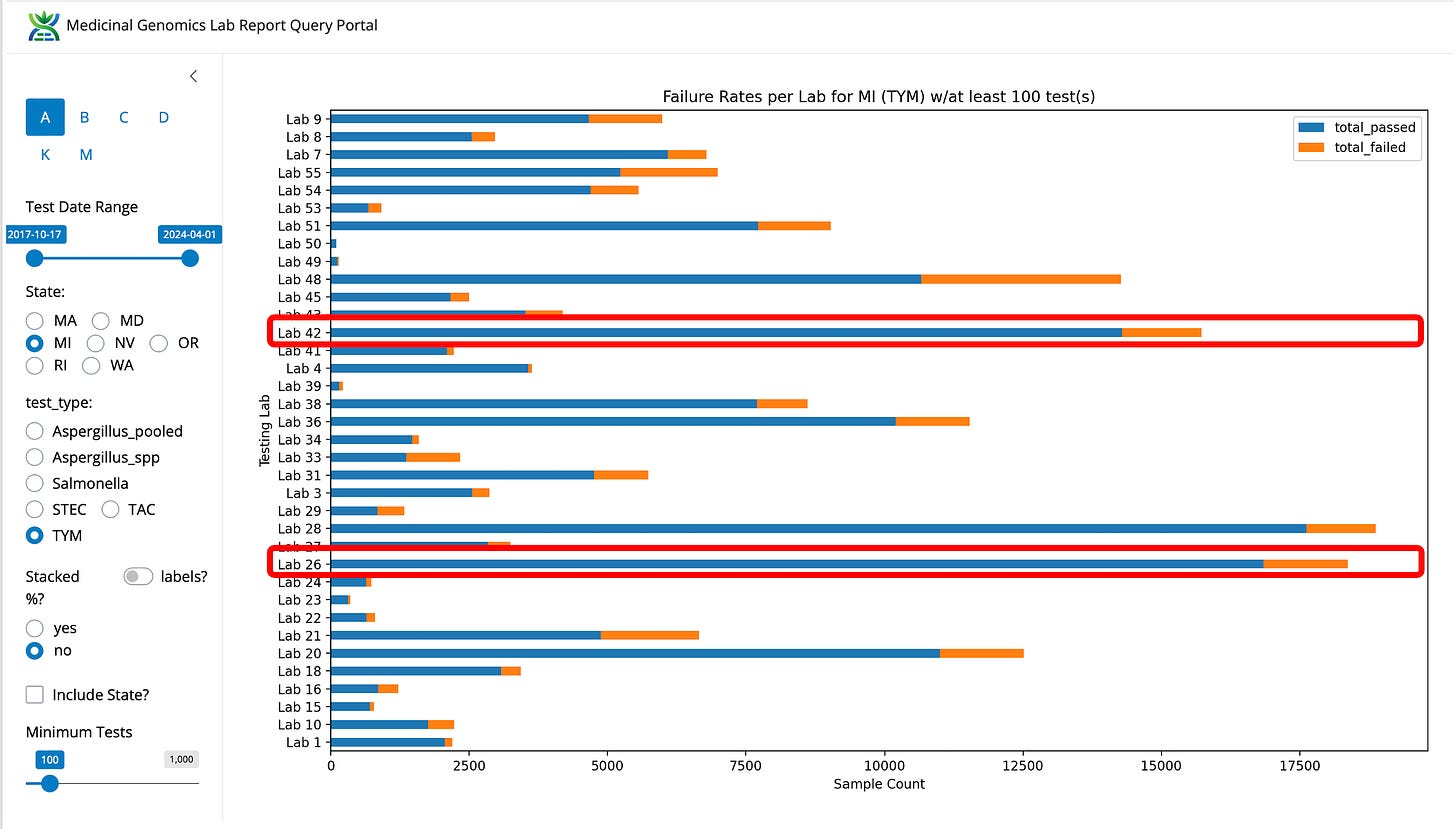
Figure 2. TYM fail rates for the State of Michigan by Lab (not normalized).
The MI-CRA also accused Viridis of inflating THC values.
Viridis developed their own THC quant method and had it certified by AOAC. Its commendable that the lab did this as few labs have put their methods through this rigorous process. Its expensive and time consuming and demonstrates rigor and good faith by the lab. But the process is not fail safe as you will see.
This certification was used to push back on the regulators questioning their THC values. Over 80% of the flower samples that were over 30% THC were from Viridis. This is an outlier result the state had to act on. This triggered a deep investigation of the Viridis AOAC approved method and triggered many letters to be sent into to AOAC to warn them of mathematical errors in the method. AOAC eventually withdrew their certification of the method.
A 2nd issue arose during a CRA audit. Sticky notes were found on instruments suggesting customized THC protocols for certain customers. Viridis was using ceramic balls to homogenize samples and they argued that the trichomes that adhered to these balls should be rinsed into the final test. This would increase THC quants but seemed reasonable on its face as those trichomes came off the flowers originally. But this doesn’t excuse different protocols for different clients. I don’t believe this accusation has resolved itself in court so lets not let reddit posts be the judge, jury and executioner yet.
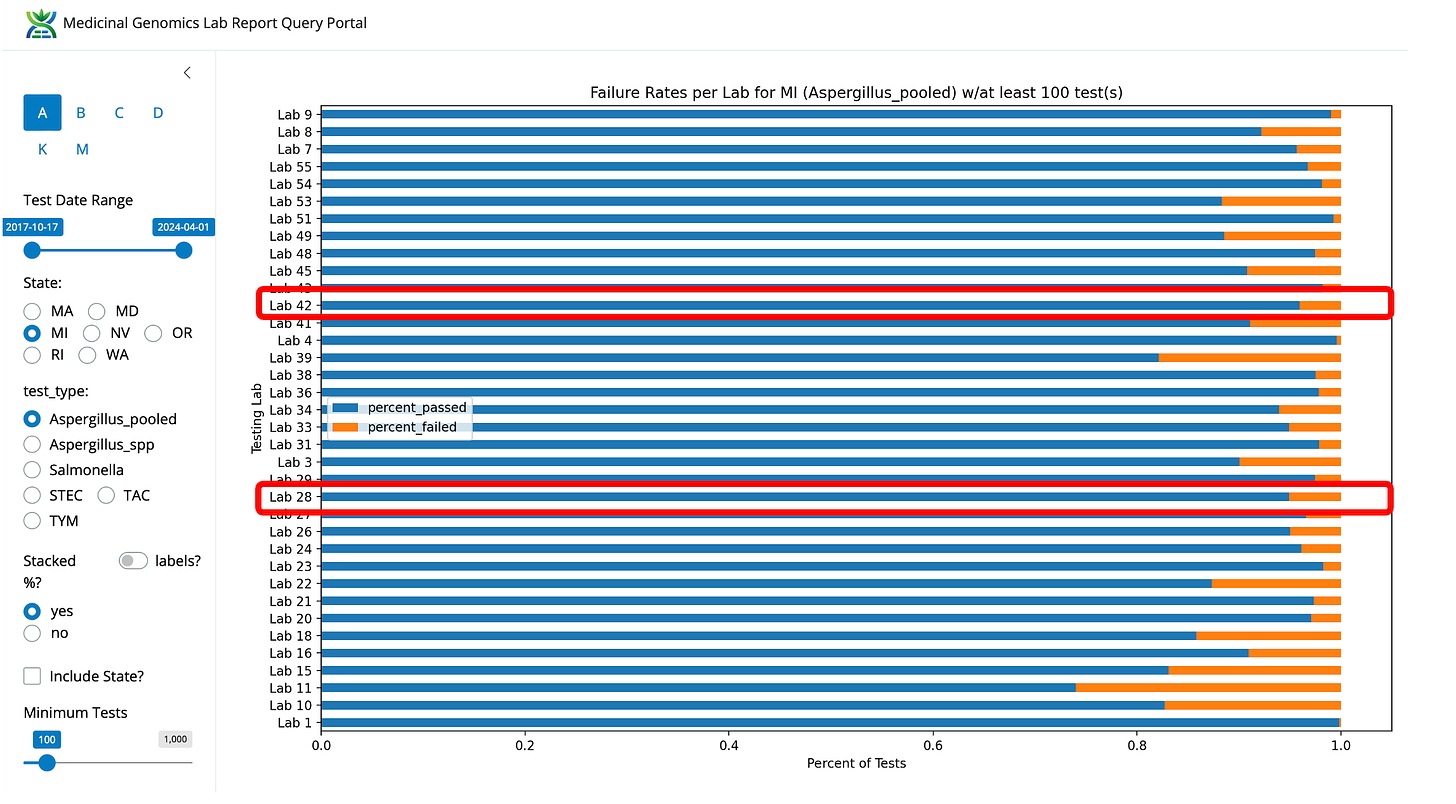
The MI-CRA lawsuit was focused on Aspergillus failure rates (not TYM). In this case these labs do have low Aspergillus failure rates but they are not the lowest in the state. Their 2 labs combined are likely the largest volume of testing in the state. This does highlight a blind spot in AOAC certifications; they do not have LODs on microbial tests.
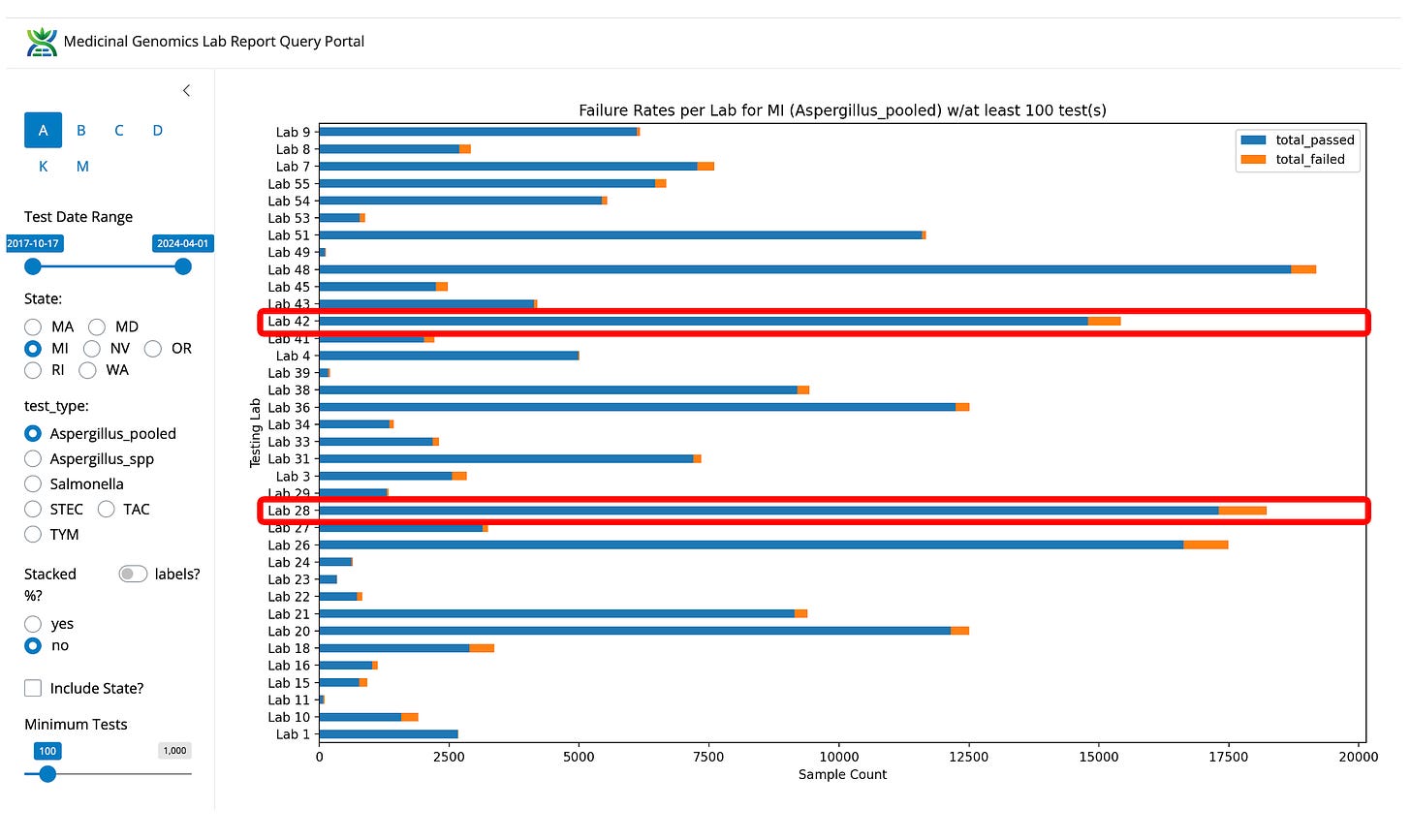
Figure 3- Aspergillus fail rates in Michigan normalized by lab. Viridis is not the lowest fail rate in the state but they are the highest volume and low fail rate lab.
Figure 4- Aspergillus fail rates in Michigan by lab (not normalized). Viridis is not the lowest fail rate in the state but they are the highest volume and low fail rate lab.
Inclusion and Exclusion Testing
A second blind spot in the AOAC process is the Inclusion and Exclusion testing. Manufacturers can source these organisms from their own biobanks and some providers do a lot of this. At Medicinal Genomics we only source organisms from Biobanks that provide part numbers other labs can order and verify our work.
If you source these organisms from your own private biobank, no one else can order these organisms to verify the claims. In addition, some AOAC certificates in the Cannabis space have ATCC part numbers (ATCC is a well respected BioBank) that are expired suggesting these companies just copy and pasted data from previous AOAC certificates not bothering to realize many of the organisms they list are expired and no longer available.
So what was all this change and why did qPCR get the short end of the stick in Michigan? qPCR was only ‘banned’ for enumeration tests like TYM. It was never banned from presence absence tests like E.coli, Salmonella and Aspergillus. In fact a ban is the wrong term. Michigan demanded all tests have AOAC approval and at the time AOAC didn’t have an SMPR for TYM in cannabis. The E.coli, Salmonella and Aspergillus SMPRs were published and underway with many providers. This lead to the AOAC setting up a TYM specific Emergency Response Validation (ERV) for Michigan Cannabis markets.
Michigan started with a 10,000 CFU/g TYM action limit which moved to 100,000 CFU/g TYM action limit around Feb-March of 2021. Around August of 2021, the lab switched methods to something that looks more culture based as the dynamic range narrowed. This is likely related to the Emergency AOAC study that was performed in the state that used a culture based system as a measurement of the truth set in Michigan.
Figure 5A- TYM data from Viridis. You can see the regs changed multiple times in 2021.
Figure 5B- TYM data from Viridis. You can see the regs changed multiple times in 2021.
You can actually see this TYM migration from qPCR to Culture in the Michigan data.
After the PCR|Culture line, you can see a cap on the measurements at 1M CFUs and striated data points at 10^1, 10^2 CFU/g (labeled Dilution factor). These are the result of dilution factors in culture based systems. Since plates can only fit ~50 colonies before they get too numerous to count (TNTC), you must plate multiple dilution of the sample to survey several LOG scales of microbes. PCR is known to have a linear response over 6 orders of magnitude so fewer dilutions are required to span 1 million fold difference in counts. So plating has a narrower dynamic range than qPCR.
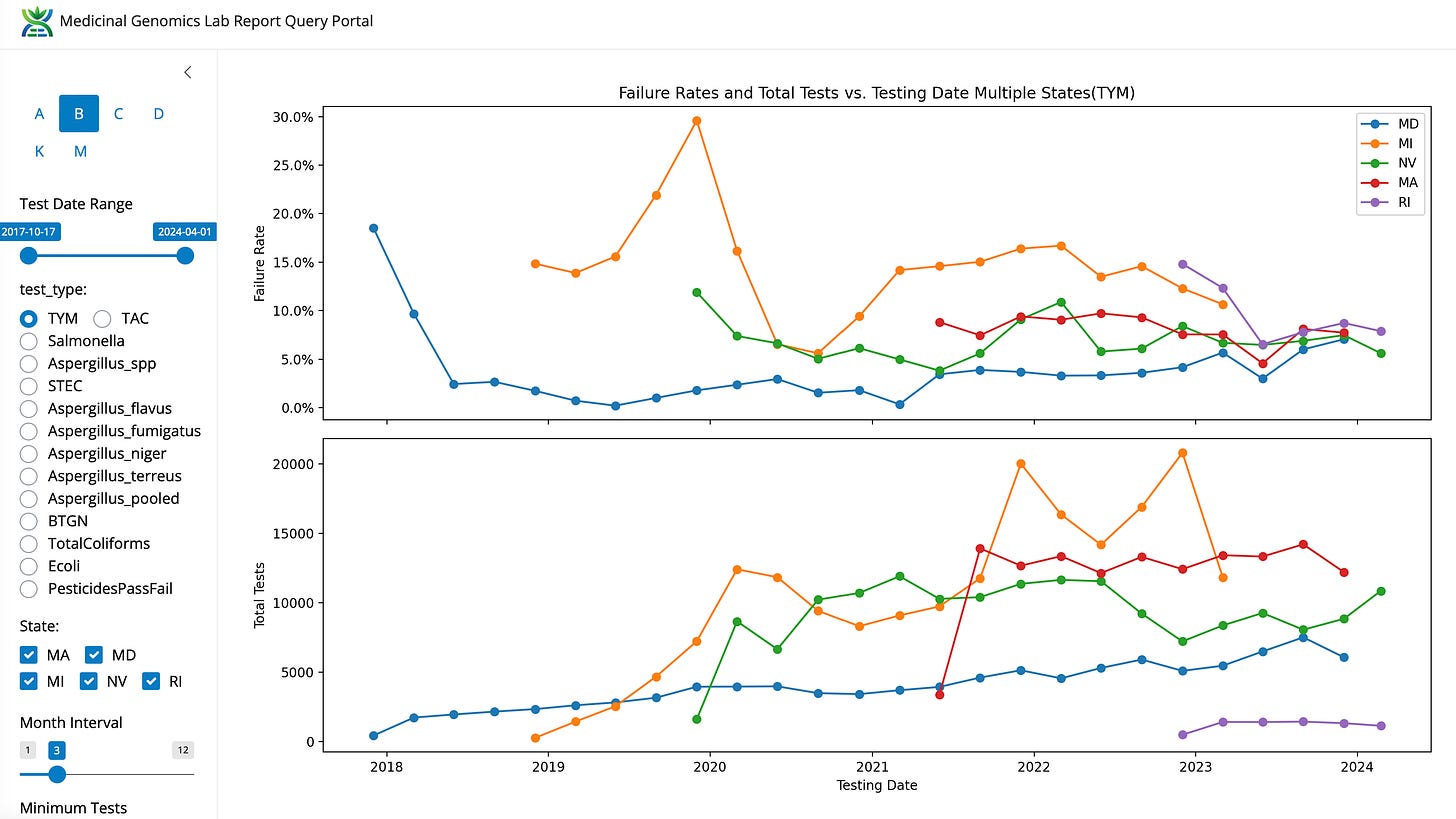
This data needs to be taken into context. Michigan (Orange) has one of the higher TYM failure rates compared to 4 other states. You can also see the fail rate drop from 30% down to 13% once the 100K TYM limit was put into place.
Figure 6. TYM and mold fail rates per state. Both MD and MI changed their recreational market TYM action limit from 10K to 100K CFU/g at different times.
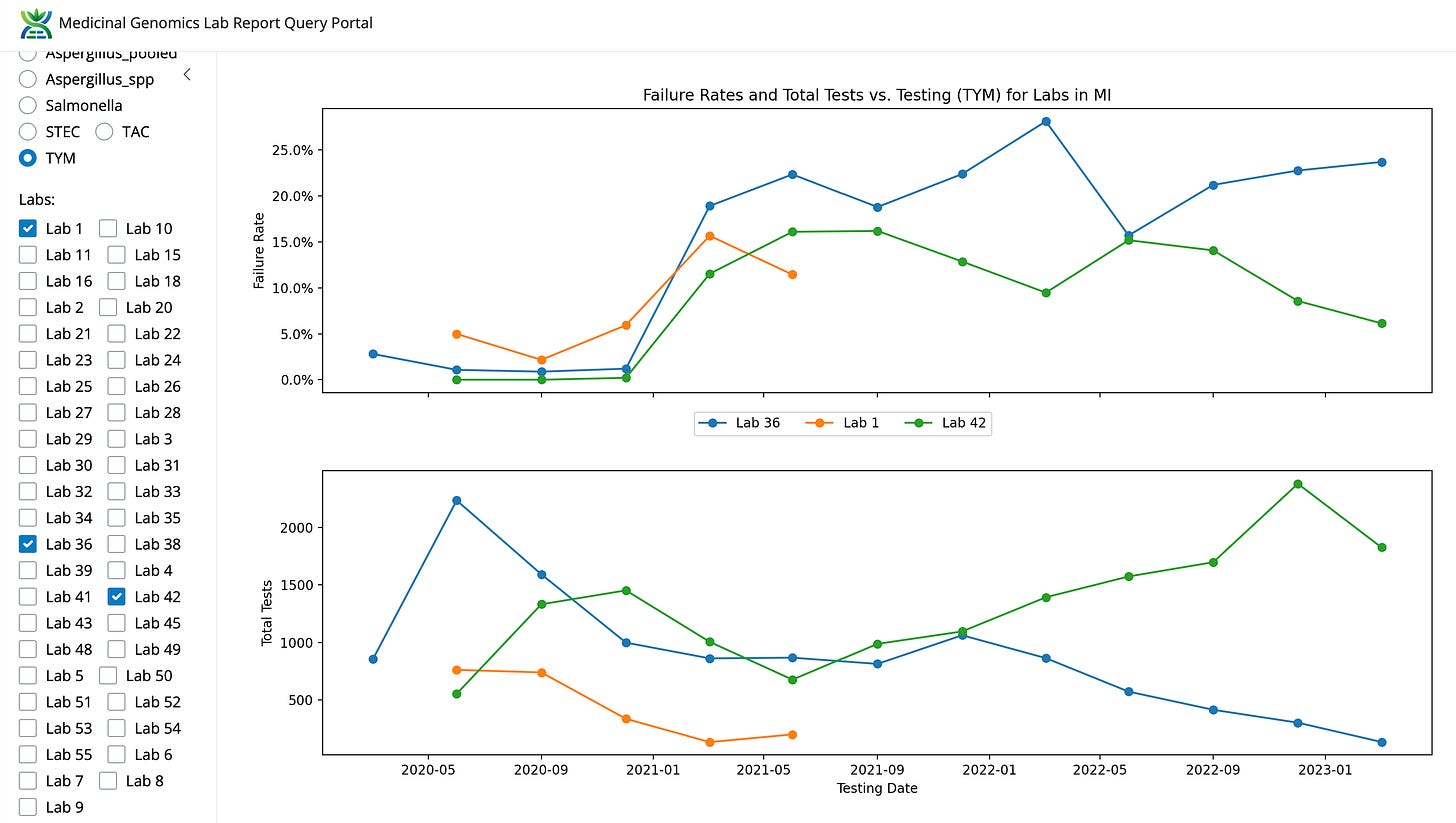
You can also see that the labs with the lowest TYM fail rate soak up all the volume in the state. This may be a tricky thing to police when the regulators are being personally sued for attempting to police it. This occurred before the Oregon lawsuit and may have weighed on the OR regulators when they were considering how to police the grows.
Figure 7. Fail rates for TYM over time in Michigan.
More recently a molecular platform was approved for semi-quantitative TYM testing using a microarray readout of an End Point PCR system. This system is being used in a Massachusetts labs that has a very odd ceiling at the action 10K CFU/g action limit (Figure 9). We highlighted this in our Oregon post.
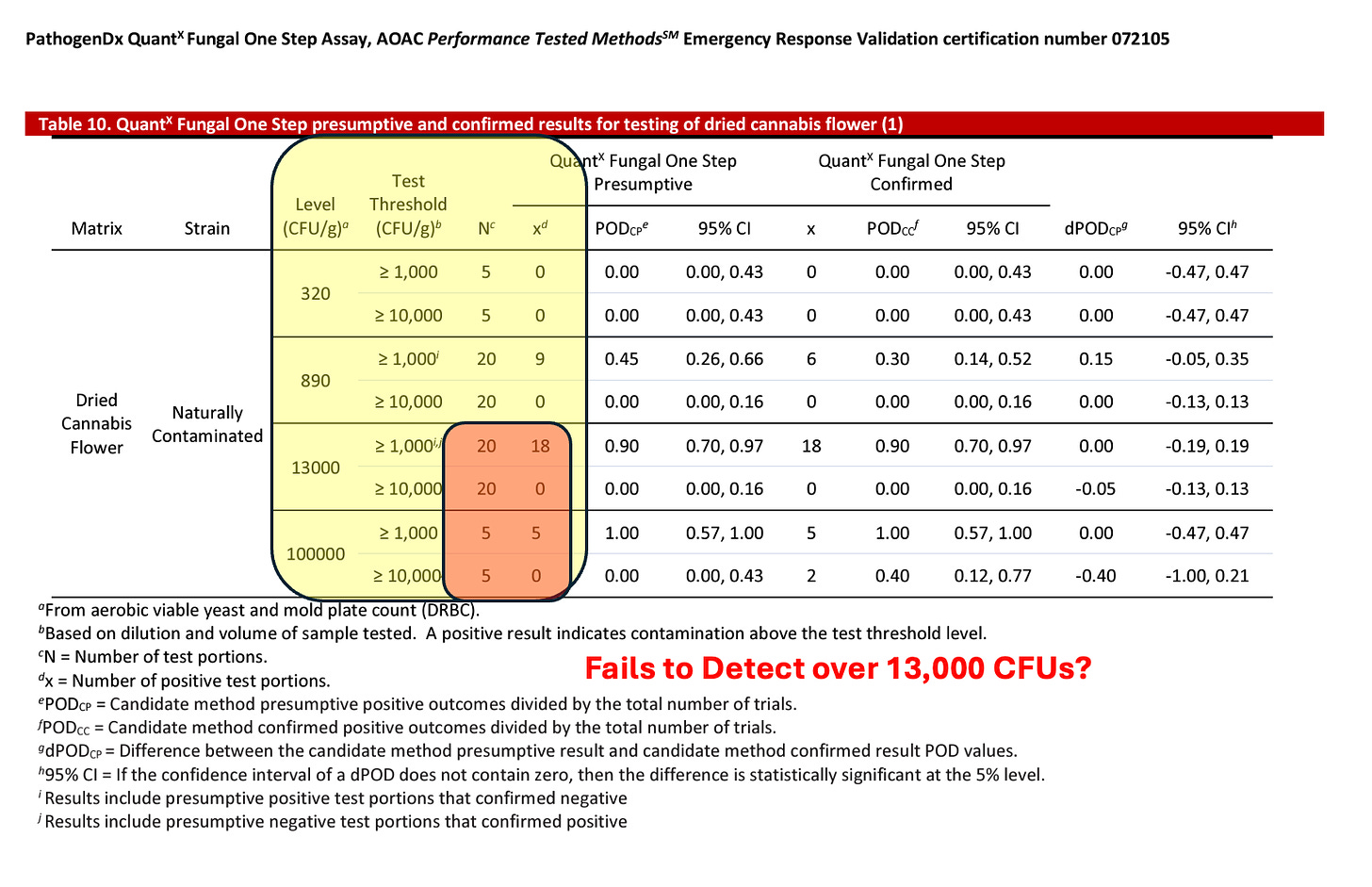
The AOAC certification for this platform appears to confirm it cannot detect over 13,000 CFUs/g
Figure 8. AOAC certification for an End Point PCR platform that shows no detection over 13,000 CFU/g.
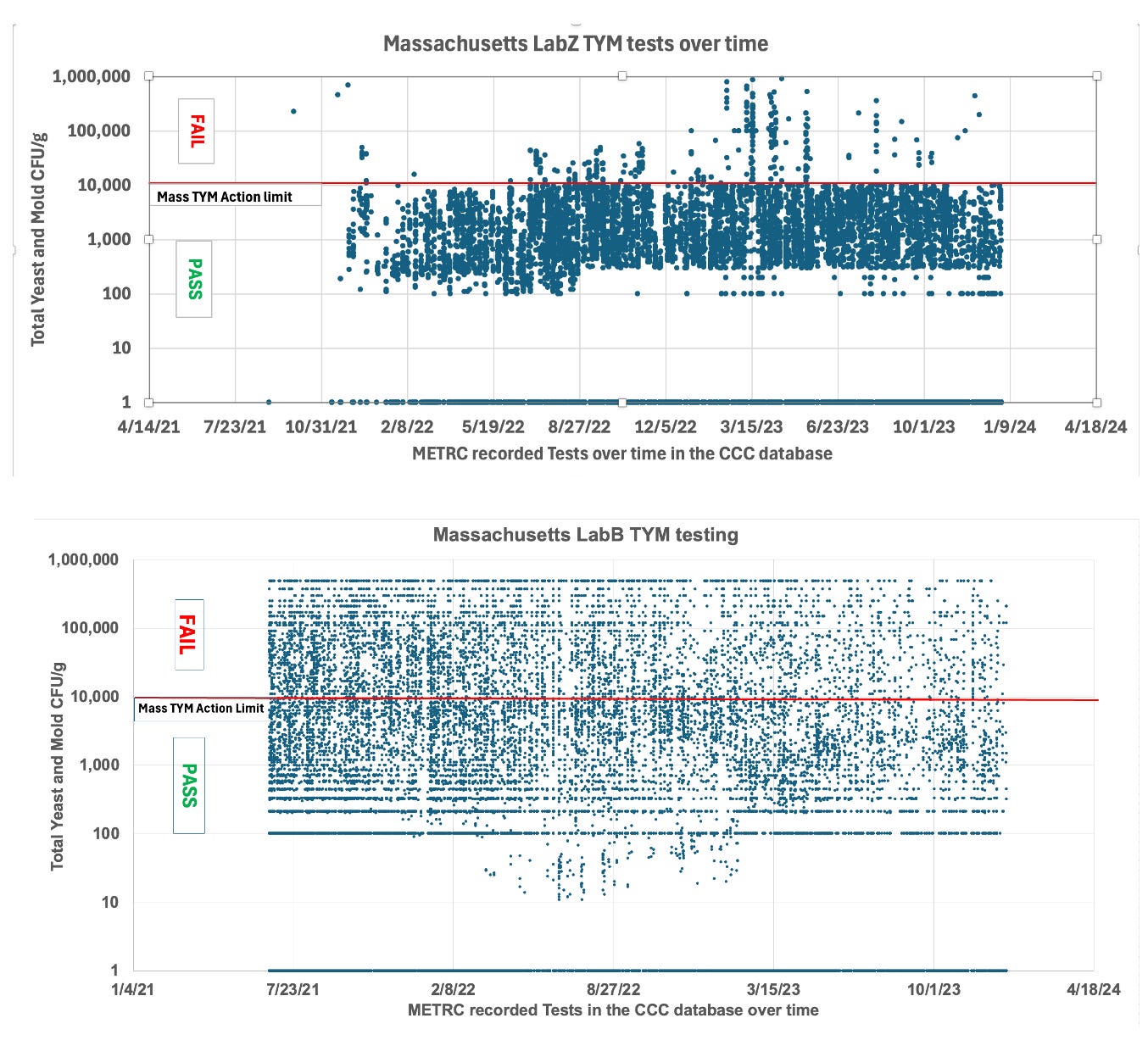
LabZ in Massachusetts is running this platform and their data lines up with this AOAC certification. LabB is running a culture based system predominantly with some qPCR.
Figure 9. TYM data from Massachusetts from LabZ and LabB. LabZ shows an unnatural distribution of CFU/g.
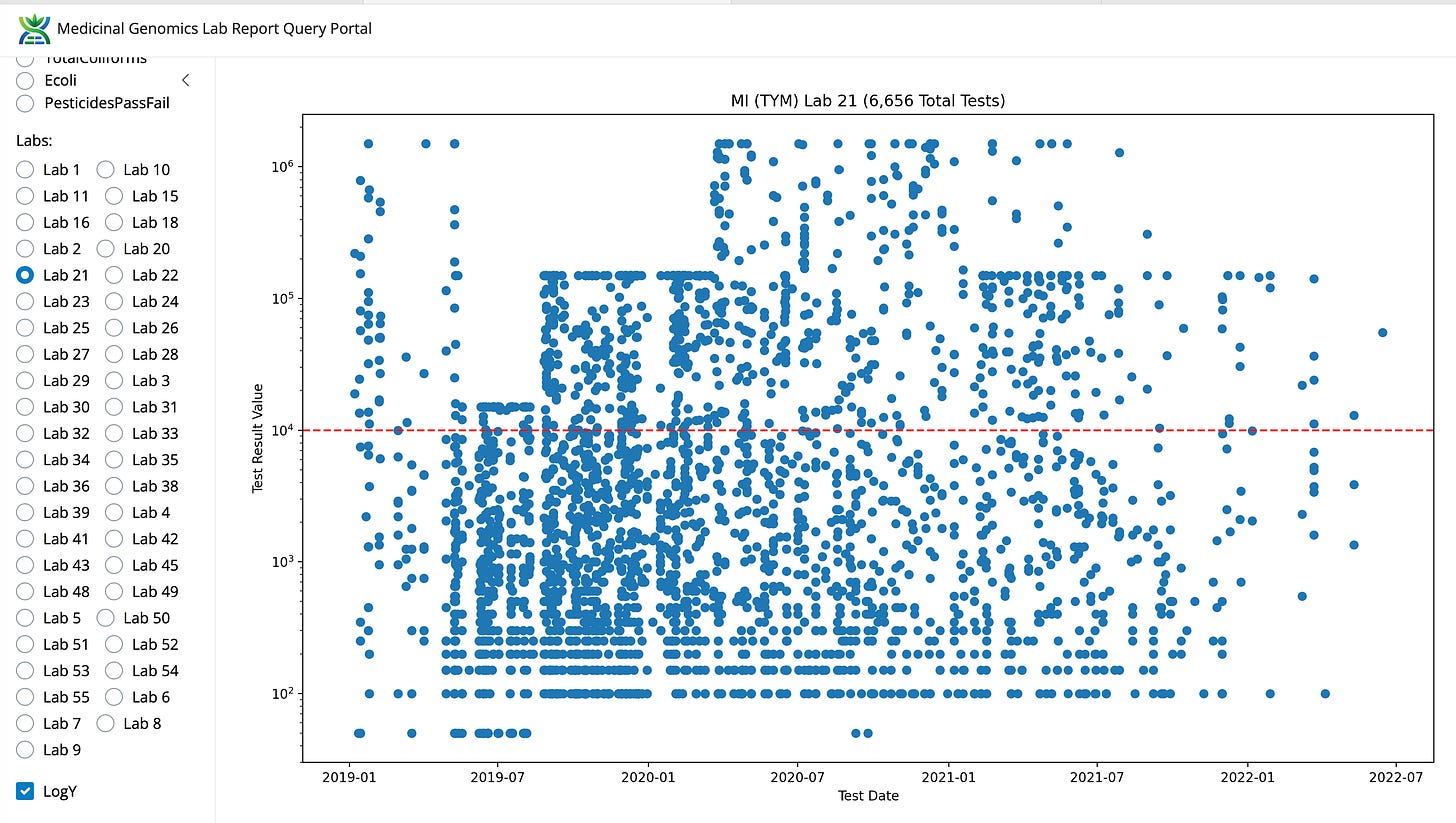
What is bizarre is that some labs in Michigan have these data ceilings move as the regs change. Its almost as if all the microbes in Michigan were on the email regarding the new action limit. This lab is no longer in business in Michigan.
Figure 10. Adjustment to the dynamic range of TYM testing for Lab21 in Michigan.
More detail on the AOAC ERV
We participated in the Michigan AOAC ERV study. The study was organized by Steadfast labs who ran a Neogen platform (Lansing MI) to measure all of the samples before being shipped to many labs participating in the study. There was High, Medium and Low CFU cannabis in the study. Its not clear how these test samples were made but we suspect a large batch of cannabis was homogenized and split into 3 batches where 2 of the batches were remediated to varying degrees to generate Medium and Low CFU samples.
Depending on how this step was performed (freezing, ozone, microwave, H2O2 etc), you can kill the CFU viability but not kill its DNA and get qPCR systems to over call the samples. Steadfast believes these dead microbes are harmless. Many other microbiologists disagree as the dead microbes are a sign that mycotoxins and endotoxins are present (Eidem et al).
A must see video on Bioaerosols from Tess Eidem.
This is in fact what happened in our testing. We failed too many samples in the AOAC study and State moved on with a culture based approach.
This was a bit surprising to us as the Emergency Response was deemed an Emergency as some labs using plating were accusing the labs using qPCR as under calling the samples. The exact opposite occurred in our AOAC study and they went with plating tools anyway! I suspect this will be material in the Viridis vs MI-CRA lawsuit.
This felt like a cheat as they measured the truth set using culture and not surprisingly the culture based system had better agreement with culture. This is particularly true if you over homogenize, freeze or ozone treat/irradiate samples. These steps are not always present in normal cannabis samples and are less required in states with 100,000 CFU TYM action limits. Illinois is a better state to look at for remediated samples as their requirements are so low that many more people use remediation tools.
Why do qPCR and Culture give different pictures?
In our previous Oregon post, we already touched on how only 5% of microbes can be cultured (some species don’t grow on your selected carbon source) and how qPCR is a more inclusive technology as it simply depends on if your organism has DNA (they all do) rather than if you selected the right carbon source to capture their growth. We published a paper critiquing the design of this plating-favored study citing many artifacts in the study design related to plating failing to pick up the endophytic fungi (fungi inside the plant) and the culture carbon source creating a narrow survey of the fungi on the plant. We even performed whole genome sequencing on all of the colonies on different plates to demonstrate how many off target bacteria they detected.
These differences are usually simply addressed by benchmarking your qPCR signal to the counts you find on plates. This benchmarking is very sensitive to which carbon source you plates use to grow the microbes. Chang the carbon source and vastly different microbes grow. Once calibrated to a given plate type, this creates a CT to CFU conversion equation. Putting this equation public for others to scrutinize is critical and in some States like Florida we know of regulators that confirm the labs are using the right CT→CFU/g equation for a given assay. Some providers don’t provide these equations and you must upload your data for them to analyze. This is a risk as it gives labs plausible deniability of any undercount as they defray that liability to the kit providers black box in the cloud.
What about Viability?
Plating only detects the epiphytic microbes (microbes that grow on the outside of the plant) that can grow on one particular carbon source and there is no guarantee these microbes will then also grow in a human host as the human host has different carbon sources and an immune system. Nevertheless one can easily plate many samples and qPCR these same samples with ITS primers and correlate the qPCR signals with the CFUs found to build a predictive equation.
Enrichment and Live-Dead discrimination
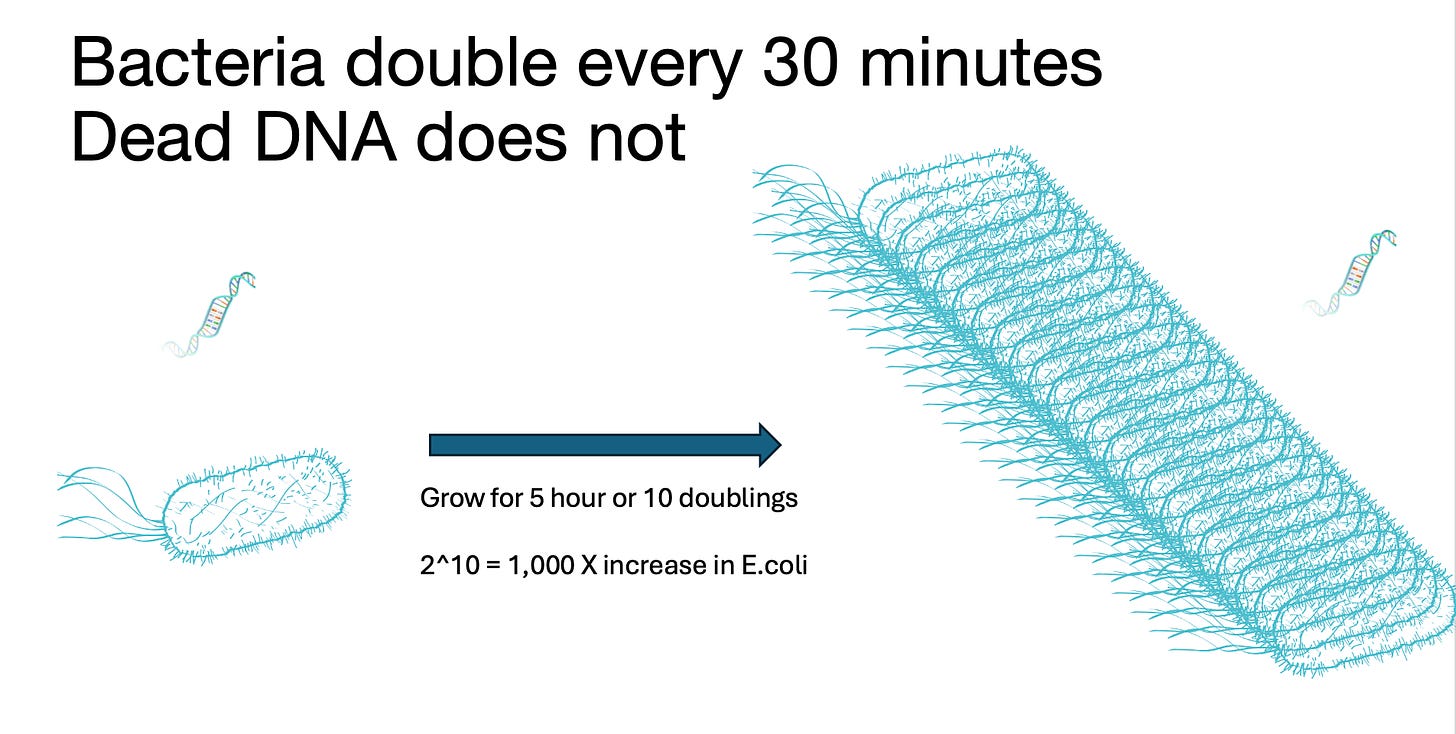
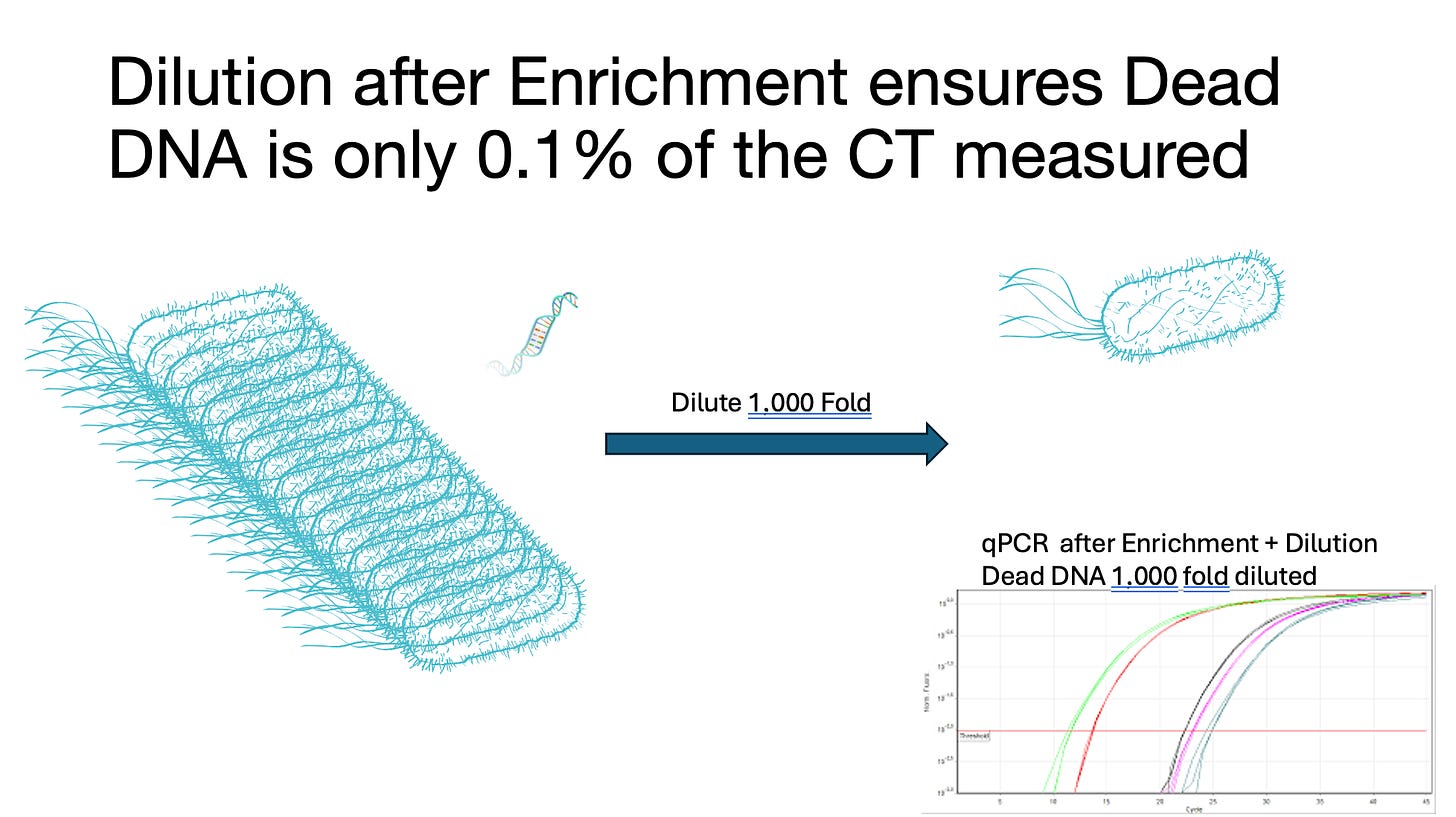
This qPCR →Plating correlation works well until you have millions of microbes present that you partially kill 99% of. Now you have only 1% of the microbes that can form colonies but all of them that can qPCR.
As a result, qPCR systems need an enrichment or a live-dead discrimination tool when used with irradiated products. Some methods simply grow the samples for 10 doublings (1000 fold more microbes) and then dilute the sample 1000 fold to dilute out the dead DNA before qPCR. The viable microbes replicate and the DNA stays at the same concentration or is consumed by the microbes under growth. This is termed Enrichment and in some states is mandated on tests that have low action limits. Our Aspergillus tests have an Enrichment process built into them as they are low action limit tests. at 10,000 CFU/g and 100,000 CFU/g Enrichment can be replaced with Grim Reefer (DNase method) to save time.
Figure 11. Enrichment process visualized.
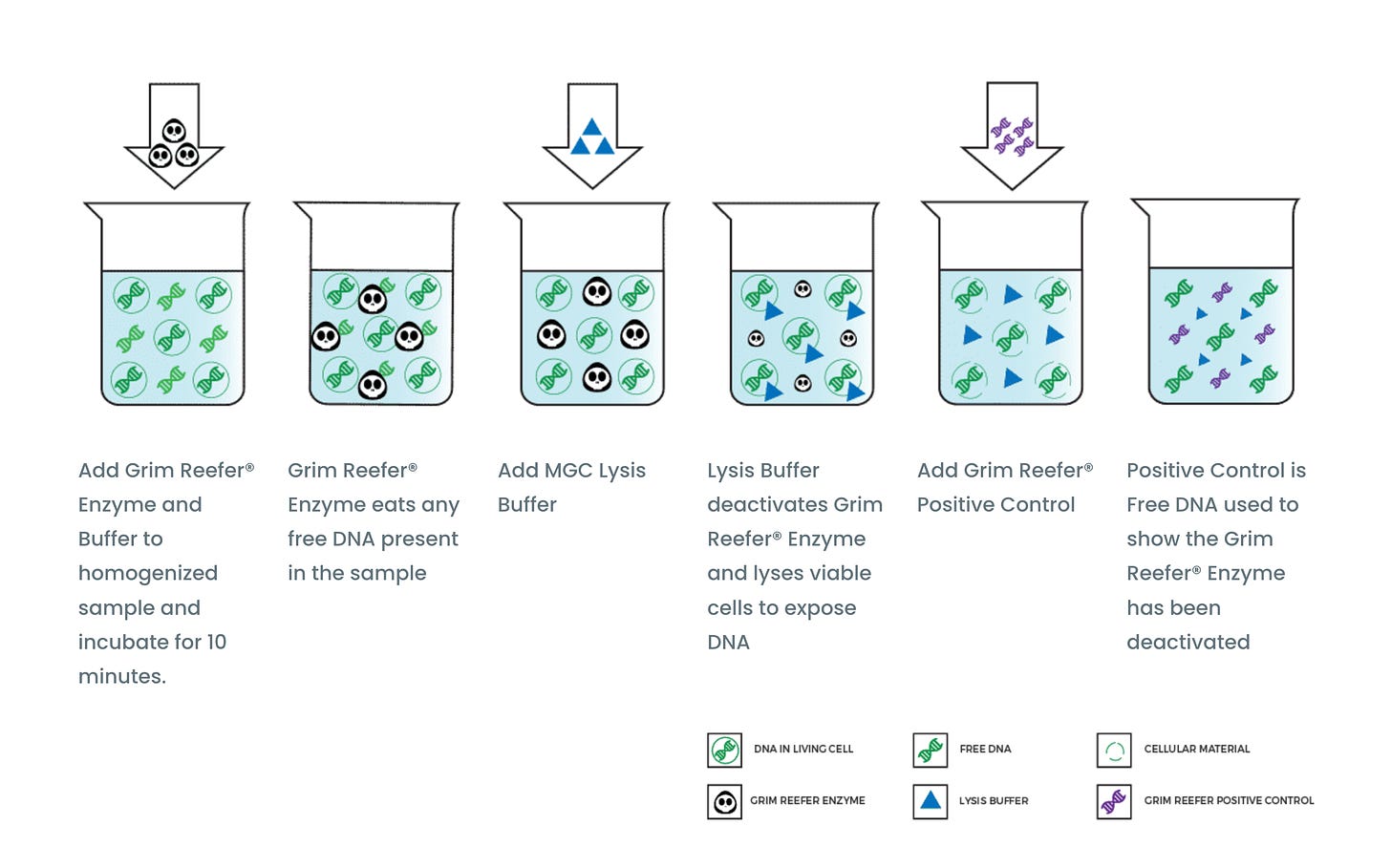
Tests with higher action limits and longer doubling times (Fungi double every 2-3 hrs while bacteria double every 30 minutes), a faster approach is to use a DNAse that erases DNA outside of a cell membrane or cell wall. When DNases are used, you must be careful to kill the DNase or it will also destroy your PCR primers.
Some methods use heat to kill this DNAase. 95C for 10 minutes is common but this temperature also lyses open cells and now you have a race to kill the DNase before it starts eliminating DNA from cell that were living before you tried to heat kill the DNase. 10 minutes is too long of kill time for an enzyme as cells lyse open immediately at 95C, so we screened many small molecules that could destroy the DNase instantly. We filed and were awarded a patent on this insta-kill method as it was more true to plating.
Figure 12. Grim Reefer process visualized
Some AOAC certified tests use the heat kill method and their AOAC certifications show evidence of the Free DNA removal steps eliminating viable colonies.
This topic was also covered on Steadfasts website.
After the Michigan ERV, we recalibrated our test to the DRBC carbon source MI-CRA chose as the reference standard. Its important to note that they made this change late in the study and several molecular based platforms were caught off guard by this last minute change. We had our qPCR system benchmarked to PDA listed in the FDA BAM. This last minute change to a carbon source that grows fewer microbes meant that our platform would overcall the study.
Despite this, we now have the platform benchmarked to DRBC and it now has AOAC approval using this DNA removal step and it is the first quantitative qPCR test ever approved for TYM. Other semi-quant methods are also now AOAC approved but Medicinal Genomics has the only one that is fully quantitative. This cost us significant financial harm and market share in Michigan.
This statement on Steadfasts website is now out of date.
Even with this Live-Dead removal (termed Grim Reefer for MGC and Free DNA Removal for other platforms) qPCR tends to get slightly higher counts for heavily irradiated samples (See Jini Glaros talk at CannMed 2023).
Many argue the qPCR methods are a better safety signal, as heavily irradiated samples are simply counterfeiting the plating based methods but the irradiation does nothing to remove the mycotoxins or endotoxins that still exist on those products and it is not required to label remediated products. The point of these total tests is to measure the bioburden of a sample (or the cell viability), however this bioburden currently does not have to consist of cells that can culture in human tissues. Presumably it is meant as a proxy for the bioaerosols that might be present with inhalation based product. With lab shopping, many grows will send irradiated samples to labs known to plate and everything else to qPCR shops that can get a faster answer.
Conclusions
In summary, microbial testing is a complicated space. There is evidence of lab shopping in the Michigan data in that the labs with the lowest failure rates tend to grow the fastest and these labs are also ‘accused’ of THC inflation by the MI-CRA. Some policing of this is in order but the arguments of over who is right and who is wrong may remain controversial as the MI-CRA doesn’t have its own testing lab. It must rely on Viridis’s licensed competitors to spot check Viridis and this remains a conflict in the trial.
Likewise, when the MI-CRA spot checks samples it can be done months later which begs the question if the sample was accurately tested when it left the lab but grew on the shelf.
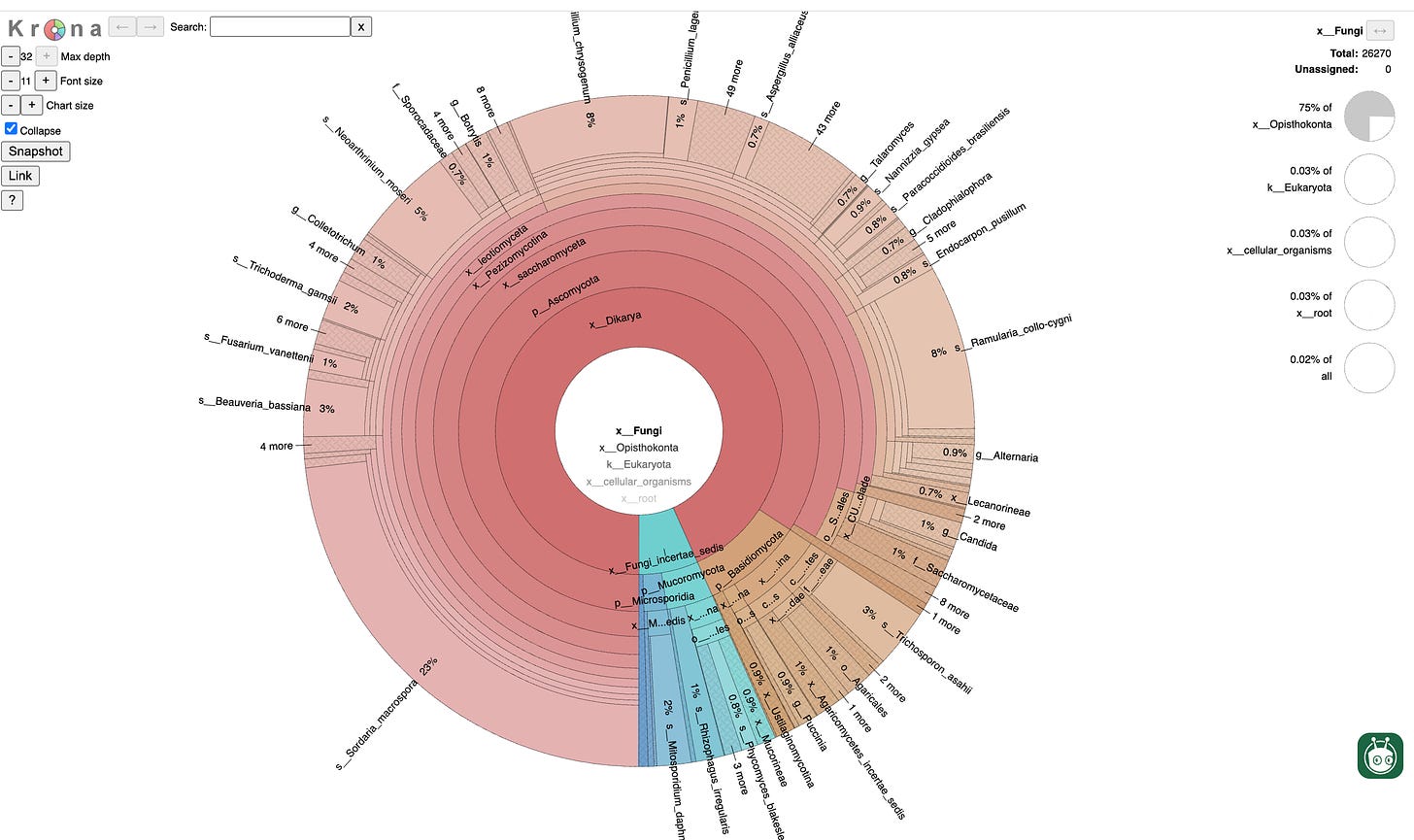
Another factor in any recall consideration is that the state should run all platforms. To pick one vendor as the truth set enables regulatory capture. Every lab will be subtly forced to switch to the vendor the state uses or face potential discordances, destroying competition in the market place. This is currently creating chaos in California as recalls issue. California is not transparent on what methods they use to determine recall status and it is well known from some vendors own literature that some kits in the field have false positives for Aspergillus tamarii. At Medicinal Genomics we attempt to settle these debates with whole genome sequencing. We have the largest cannabis microbiome database in the world and can compare a given sample to everything ever seen before and help with precise speciation of a questionable qPCR call.
Figure 13. Fungal microbiome report from whole genome sequencing of sample Purple Kush-St-Flower from Zamir Punja. There are 70 Cannabis whole genome sequences which include their Bacterial, Viral and Fungal microbiome on Kannapedia.net
When regulators attempted to police the Michigan Aspergillus recall, they may have overstepped their authority? The trial is still under way so its too early to conclude this. I’m not a legal expert but my sense is this could have been policed more actively before $229M of product built up for recall. There were prior smaller enforcement actions in Michigan. Just none that amounted to 60% of the market under recall.
Once $229M was on the table, lawyers were mobilized. Smaller, more frequent corrections would have been easier for businesses to absorb and adapt accordingly. This is an argument for public data as the discordances may be picked up by the public in more real time.
Constantly changing the regulations is one way to weed out small labs. This also happened in MD. Smaller labs can’t afford to keep up with all the revalidation costs so these changes should be vetted with transparent public comment periods and the frequency of change should be a consideration given many of these labs have to operate without banking or the capacity to write off their business expenses according to 280E regulations.
Had the data been transparent and public, outlier data could be flagged in real time and policing could have occurred more incrementally. Labs have a good argument here. Without transparent data, a lab has no idea they are the lowest failure lab. Transparent data acts like as a continuous Proficiency Test on the field and allows consumers to chose if they want cannabis that was tested at such a facility. This also enables the regulators to leverage many consumer advocate analysts who can help them identify trends in the data. Not every consumer is the same. Some consumers have been consuming untested black market cannabis for years and may not want to pay for samples put through more expensive and more stringent testing. Others may be very sensitive to mold and have higher standards.
The age old debate on qPCR vs Plating never seems to end but I will conclude with 10 closing thoughts.
1)Independent of platform type, we can see labs that have abnormal data. Certain tests in Maryland and Nevada are ‘plating only’ tests and these games are still being played.
2)Plating only captures 5% of the microbes that can be cultured and often fails to survey the endophytes (microbes that grow inside of plants). In order to get microbes out of plants, you often need to lyse open plant cells and these conditions destroy the viability of microbes you are attempting to culture.
3)100% of yeast and molds have ITS DNA sequence targeted by qPCR systems. qPCR is always forced to have correlation with this blind plating tool. This naturally leads to poor correlation with plating. Some studies over call with qPCR and some under call depending on the study design. The tendency is to blame platform difference for different results but this isn’t always the case. Even when platform choice and competition is narrowed, thus raising the price of testing, the lab shopping continues.
4)Remediation tools can zero out a plating result more readily than a qPCR result.
5)Total count tests don’t measure human pathogens but are justified on a total bioburden basis. This bioburden should include the bioaerols these dead microbes have left on the plant after remediation.
6)Remediated samples are not necessarily safe just because plates fail to see the risks.
7)Lab Shopping and Lawfare will ensure this debate rages on. The solution to this isn’t for the state to pick one platform and declare it a gold standard from which they issue recalls, but instead to use Next Generation sequencing (WGS) to truly identify what is present in each sample. We have over 1,000 Cannabis microbiomes on Kannapedia.net and perform this frequently when ever a recall is in question. We have documented this in the past. Some of the Aspergillus qPCR are known to hit Aspergillus tamarii and Aspergillus luchensis. If the state uses only one platform for a recall and doesn’t confirm the recall with WGS, they may find themselves in court like the MI-CRA.
8)Transparent public data is the best solution. Let consumers decide and let labs have instant feedback on how they are performing next to their peers.
9)Regulating this space is not easy. Lots of conflicts and few standards will keep the lawyers busy for years to come. Transparency in the data will enable the market to help police the space. For many labs, this is the first glimpse of comparative data they have seen. It’s hardly fair to drop recalls on these labs when they’ve never seen their report card and the methods used by the regulators are not public for inspection.
10)Rescheduling/descheduling will only help this field. Expecting the regulated market to replace the blackmarket with 280E in place is like fighting the black market with both economic hands tied behind your back. Many cannabis companies face banking challenges. The inability for cannabis businesses to write off their business expenses, leaves many cannabis businesses painted into economic desperation. Labs can’t share samples across state lines so labs need to be brick and mortar replicated in every state. This easily eats up $5M in capital equipment and overhead for each state lab. This also makes it hard for labs to standardize tests. These same redundant cost structures inhibit grows from implementing internal testing to QC their process more frequently than just at the final packaging stage. Most other industries perform real time QC and a few smart grows are realizing it pays dividends just for HLVd testing alone. You can’t plate for HLVd and many cannabis pathogens. qPCR is the only universal platform that enables, fungi, bacteria, viruses and viroids to be surveyed.
Keep in mind, I am not a legal expert and I sympathize with both parties in this legal dispute as I can see valid arguments on both side of the disagreement. The lesson I have taken from this episode is that public data and more frequent secret shopper programs may have prevented all of this. We will have to leave it to the courts to decide.
Conflict reminder- We now have an AOAC quantitative TYM test that is specifically designed for Michigan markets. It was benchmarked to DRBC and rated quantitative (not semi-quantitative) by AOAC. It is the only molecular method to obtain this status and many labs in Massachussets have reported to us that it is true to plating. It is being evaluated in many labs in Michigan as it delivers data faster than plating and is very automation friendly, low cost and scalable. This thread is not meant to throw stones at any labs or regulators but to highlight the power of public data and how much it aids in objective peer review. We all want lab shopping to end and have labs compete on a level playing field where competition between the kit providers ensures accuracy and the best pricing.
Acknowledgements: Yasha Kahn for supplying Michigan testing data. Benjamin Warren for pointing out some anomalies in MI data. See Jamie Toths article on the risks from Aspergillus.