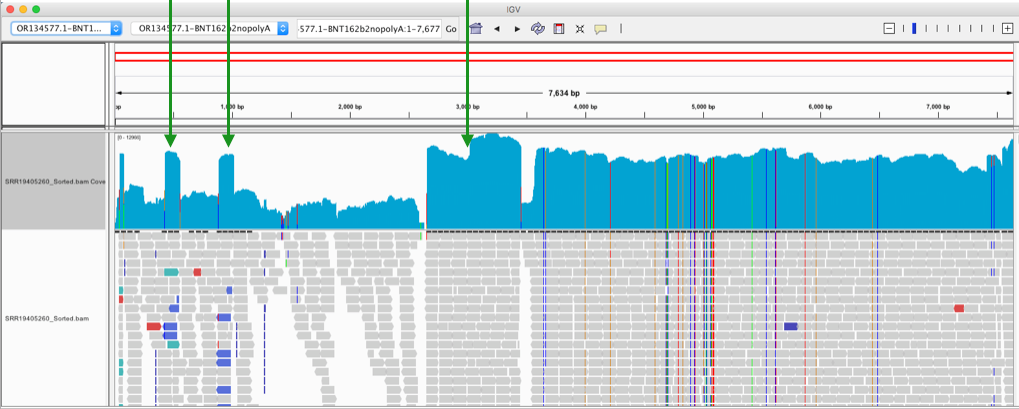
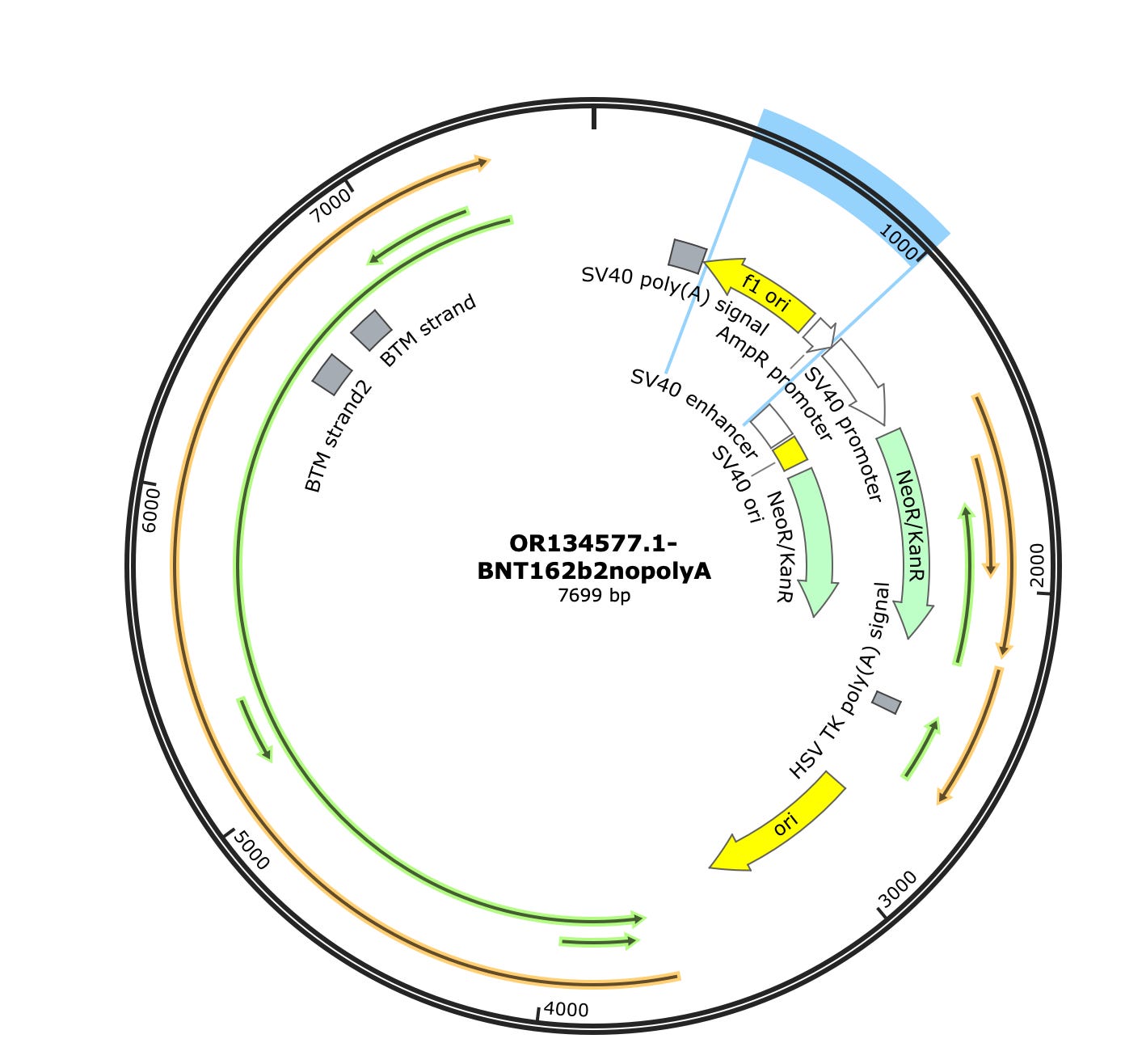
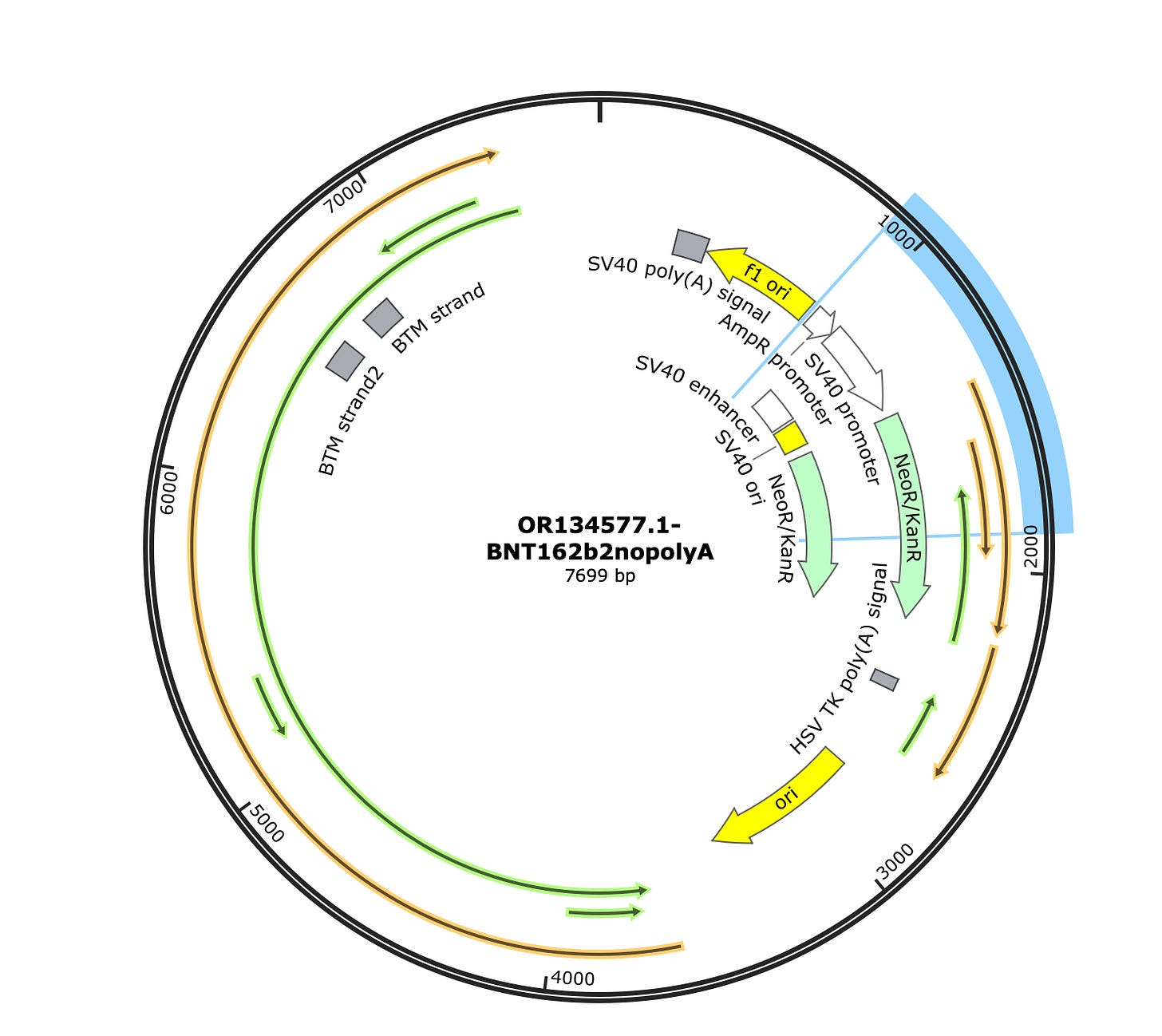
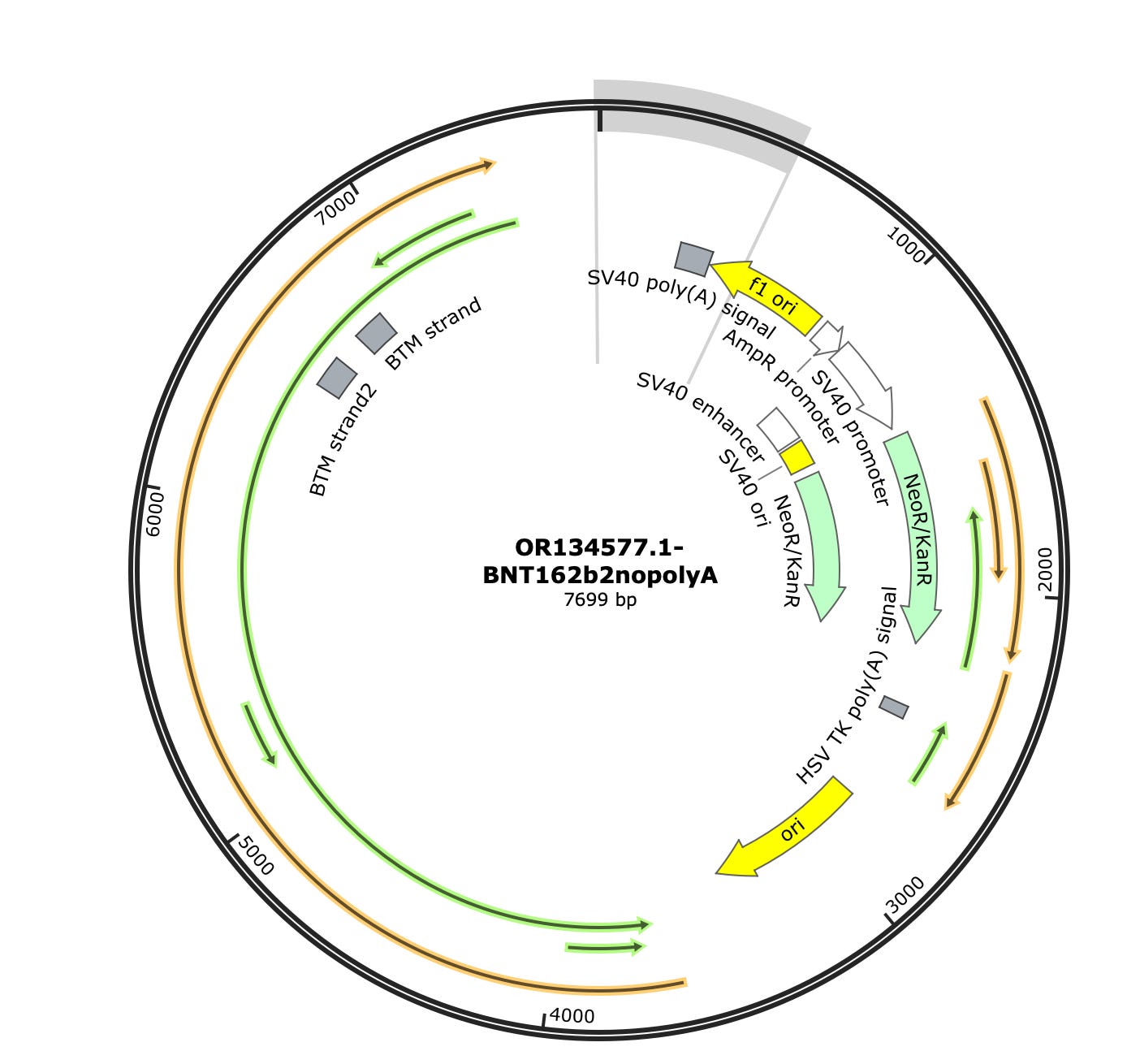
Phillip Buckhaults gave me a ring to point out some of these square towers in the vectors coverage maps (Green arrows) are likely artifacts from the pUC plasmids used to manufacture some of the enzymes in the sequencing process. The bacterial origin of replication in the middle (3,000) and F1 origin (~500) and the AmpR promoter (~900) on the left are commonly found in plasmids used to express the library construction enzymes. Their erratic jump in coverage is also an oddity. We don’t see this when we sequence the vaccine directly. These are not patient derived and if the studies sequenced a negative control, you would probably see them as well. Most enzymes used in molecular biology are expressed from a plasmid with a ColE1 ori and grown in E.coli and they share these features with Pfizer. The sequence between the Mesa’s is Pfizer vaccine.
Some of the samples (SRR19405260) do have coverage across the rest of the plasmid. There is one gap at 2630. The spike sequence which is unique to BNT162b2 is present as well, but at much higher coverage (~1500X). The SV40 promoter is around 50X coverage.
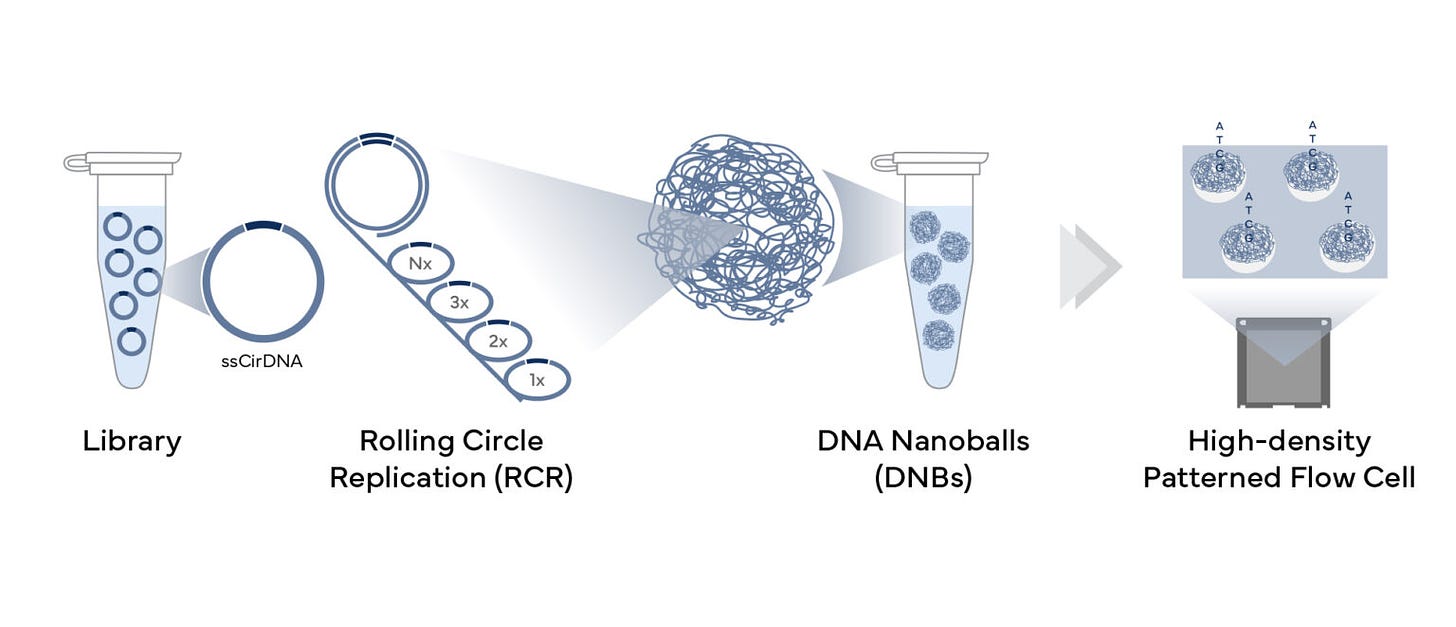
One question we had on the last post is if the 50:50 read ratio was a result of Illumina read pairing? This wasn’t run on an Illumina. The DNBSEQ G400 makes circles out of DNA and amplifies them into DNA nanoballs. This is really helpful as the index hopping drops down 100-1000 fold with this technique.
These DNA nanoballs non contain 10,000 copies of your original template. They are then easy to pack tightly into a 50nm pitch patterned array for high throughput read out of the sequencing chemistry.
Why this is important is that this nanoball based sequencer produces reads that are in a different orientation than Illumina reads.
As a result of this circulation step the reads from any given nanoball are always on the same strand. So crick stranded RNA should all be Forward-Forward reads. We have a mixture of both Forward-Forward and Reverse-Reverse reads which still implies DNA not RNA as the source of this nucleic acid.
The reads in the spike region still come back 50:50 on aligning to both strands?
If we trust samtools, this tells us these are in fact DNA generated Spike molecules and they are a little higher in coverage than the vector.
Very few of the samples are like this but the authors did respond to me claiming they did in fact use a DNA depletion step so this is a surprise anything made it through.
The crux of this being DNA is twofold.
1)Vector DNA from BNT162b2 should not be there
2)Stranded RNA-Seq should have at most 2% of reads being mis-stranded.
Argument #1 has some noise in it from what appears to be other plasmids that share a few components with Pfizer.
Argument #2 rests on Samtools getting the Read orientation correct for a sequencer that is a minority player in the Sequencing market. The US, in some act of economic jingoism actually threatened to sanction this sequencer as it was manufactured in China and had alleged ties to the PLA. I believe this threat has since been lifted.
Given the authors have responded confirming they performed a DNA depletion step, we should double check these arguments.
Phillip Buckhaults noted his CLC software disagreed on samtools strandedness analysis. This may be because samtools doesn’t anticipate the DNBSEQ G400 read pair orientation.
To address this, I took each DNBSEQ G400 read set and mapped them as singletons (no pairing).
I then counted the read strandedness as singletons in the spike and in the vector.
Using this approach the spike read look stranded. About 1% of the reads are mis-stranded which is within the rate of expectation for these methods when using pure RNA.
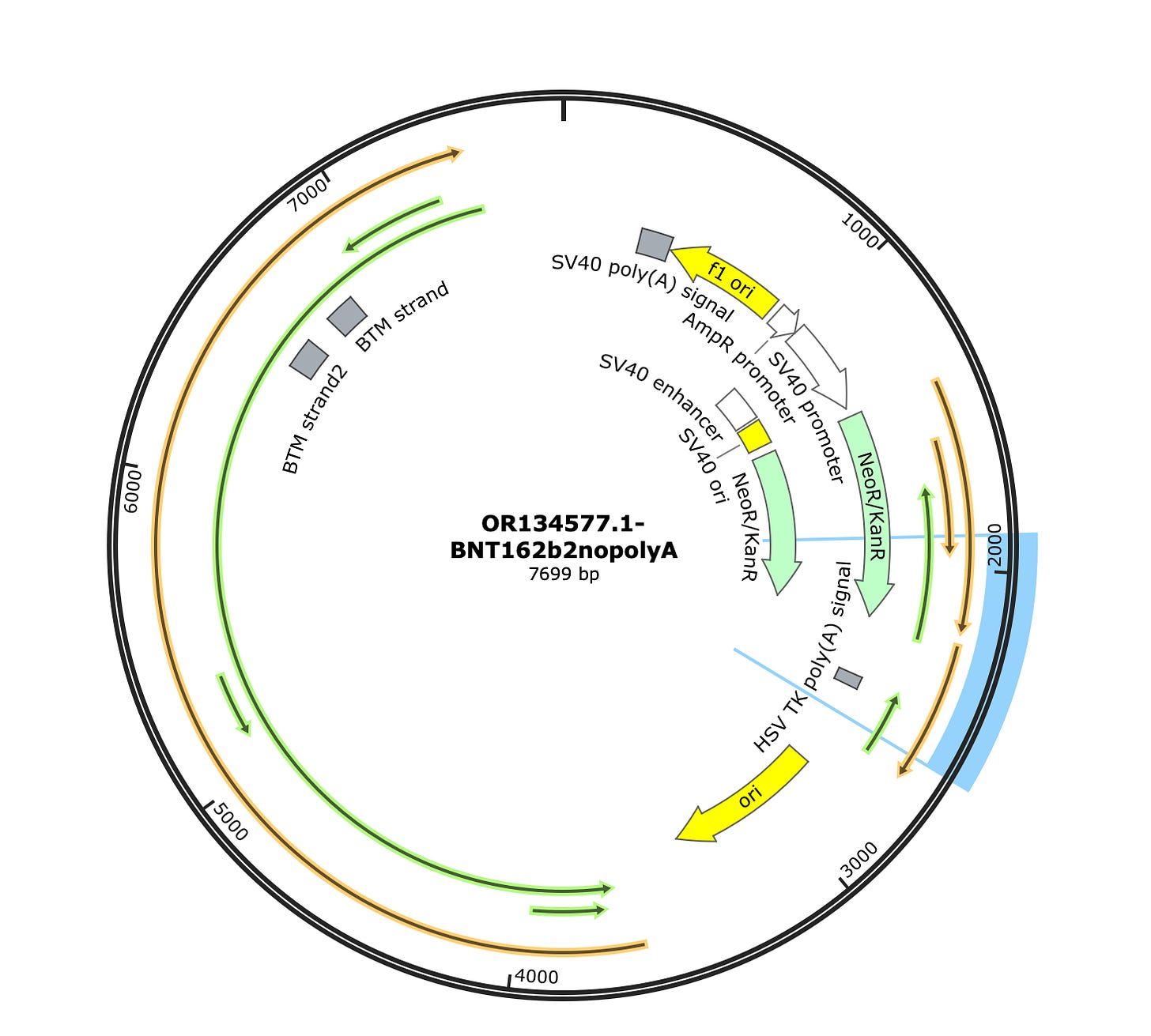
The Vector sequence I constrained to just 1000-2600 bases as this region looks unpolluted by background plasmids. A simple samtools view function can select out the reads that map to certain coordinates.
The vector reads have a much higher Watson read rate. It’s not 50:50 but its 10X over the expected background rate from these kits. Keep in mind, the authors employed steps to remove the DNA and only present RNA so we should expect much higher numbers had these precautions not taken place.
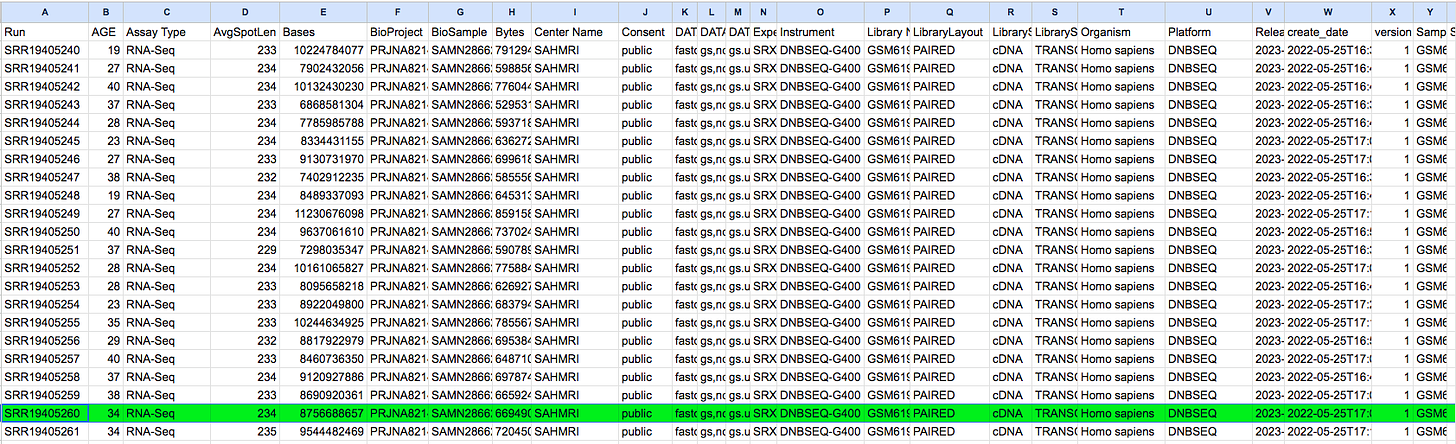
The SRA Run Table will give you details of which SRR data file pertains to which vaccinated subject. Ie was it their 1st dose and which vaccine? 34 Year old Female with BNT162b2. V2A = Second dose
To confirm this we also performed a MegaHit assembly on the RNA-Seq data for SRR19405260 and collected contigs that mapped to the Pfizer plasmid.
>k119_113047 F1 Origin of replication
TCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTGTGGTGGTTACGCGCAGCGTGACCGCTACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTTAGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGTGATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAAGGGATTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAATTTAACGCGAATTTTAACAAAATATT
>k119_16032- ColE1 Origin of replication
TGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAAACTTGGTCTGACAGTTACCAATGCTTAATCAGTGAGGCACCTATCTCAGCGATCTGTCTATTTCGTTCATCCATAGTTGCCTGACTCCCCGTCGTGTAGATAACTACGATACGGGAGGGCTTACCATCTGGCCCCAGTGCTGCAATGATACCGCGAGAACCACGCTCACCGGCTCCAGATTTATCAGCAATAAACCAGCCAGCCGGAAGGGCCGAGCGCAGAAGTGGTCCTGCAACTTTATCCGCCTCCATCCAGTCTATTAATTGTTGCCGGGAAGCTAGAGTAAGTAGTTCGCCAGTTAATAGTTTGCGCAACGTTGTTGCCATTGCTACAGGCATCGTGGTGTCACGCTCGTCGTTTGGTATGGCTTCATTCAGCTCCGGTTCCCAACGATCAAGGCGAGTTACATGATCCCCCATGTTGTGCAAAAAAGCGGTTAGCTCCTTCGGTCCTCCGATCGTTGTCAGAAGTAAGTTGGCCGCAGTGTTATCACTCATGGTTATGGCAGCACTGCATAATTCTCTTACTGTCATGCCATCCGTAAGATGCTTTTCTGTGACTGGTGAGTACTCAACCAAGTCATTCTGAGAATAGTGTATGCGGCGACCGAGTTGCTCTTGCCCGGCGTCAATACGGGATAATACCGCGCCACATAGCAGAACTTTAAAAGTGCTCATCATTGGAAAACGTTCTTCGGGGCGAAAACTCTCAAGGATCTTACCGCTGTTGAGATCCAGTTCGATGTAACCCACTCGTGCACCCAACTGATCTTCAGCATCTTTTACTTTCACCAGCGTTTCTGGGTGAGCAAAAACAGGAAGGCAAAATGCCGCAAAAAAGGGAATAAGGGCGACACGGAAATGTTGAATACTCATACTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTAAATTGTAAGCGTTAATATTTTGTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGACCGAGATAGGGTTGAGTGGCCGCTACAGGGCGCTCCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGTTTCGGTGCGGGCCTCTTCGCTATTACGCCAGCTGGCGAAAGGGGGATGTGCTGCAAGGCGATTAAGTTGGGTAACGCCAGGGTTTTCCCAGTCACGACGTTGTAAAACGACGGCCAGTGAGCGCGACGTAATACGACTCACTATAGGGCGAATTGGCGGAAGGCCGTCAAGGCCGCATGATTTAGGTGACACTATAGAAGAGGCAGTGGGAGTCCCGCCAGGGAGGCAGGTGACCAATGCTCTTCCGAGCTCCTGGGACCAACCACTGAGACGTCGTTGCGCTCACCGGACCCGATGCTACAAACCCGAGGTGCAGCGTTGACAGCTCGTGGATAGCCGGGCTGGAGTTGCGTGTGAGTGTAAGAGGTACCCGATTGCGAGAGCGGATCACACCCTACCGTTGCCCGATGGAACGCTGGCGCGGTCTACCGCTCGCTGAACGGCTCCGTGAACGGTACTTCCGTCCCCTTAATGTATGGGCCCACGCTGTCTTAGAGCGCGCTAAGGTGATTTGTCGAGGTGGAGGAGACGGGCGACTGCGGGAAGAGAAGTCCCTGAGCCATGTACCTTGCGGTGAGGAAGGCGCGAGGGGGACGGGCGGTTCTTCCGTACTGTGGAGGGGCCGCGCCCAATAATGGTCGTGTCTGAATGTTTACTGCGCCTCCGTAACGCGGCCGCCTCTTGACACCGCGGCTCCCTACCCGCCTCGGGCGAGTGAGCAGGTTTCAGAGAGAGTCTAACAAGAGGGTTCTCTTATCTCGCCGCAGCTCGTACAACATCCCCAGGTAACTATGTGCATCATTCTAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAGAATTCCTGGGCCTCATGGGCCTTCCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGTGCCAGCTGCATTAACATGGTCATAGC
>k119_16033- AmpR- SV40 Promoter/Enhancer- Neo/Kan
TTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCATGAGACAATAACCCTGATAAATGCTTCAATAATATTGAAAAAGGAAGAATCCTGAGGCGGAAAGAACCAGCTGTGGAATGTGTGTCAGTTAGGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCGGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAGATCGATCAAGAGACAGGATGAGGATCGTTTCGCATGATTGAACAAGATGGATTGCACGCAGGTTCTCCGGCCGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATGAACTGCAAGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGGAAGCCGGTCTTGTCGATCAGGATGATCTGGAC
>k119_162703- spike
CGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGCTCACATGTTCTTTCCTGCGTTATCCCCTGATTCTGTGGATAACCGTATTACCGCCATGCATTAGTTATTAATTAATACGACTCACTATAAGAATAAACTAGTATTCTTCTGGTCCCCACAGACTCAGAGAGAACCCGCCACCATGTTCGTGTTCCTGGTGCTGCTGCCTCTGGTGTCCAGCCAGTGTGTGAACCTGACCACCAGAACACAGCTGCCTCCAGCCTACACCAACAGCTTTACCAGAGGCGTGTACTACCCCGACAAGGTGTTCAGATCCAGCGTGCTGCACTCTACCCAGGACCTGTTCCTGCCTTTCTTCAGCAACGTGACCTGGTTCCACGCCATCCACGTGTCCGGCACCAATGGCACCAAGAGATTCGACAACCCCGTGCTGCCCTTCAACGACGGGGTGTACTTTGCCAGCACCGAGAAGTCCAACATCATCAGAGGCTGGATCTTCGGCACCACACTGGACAGCAAGACCCAGAGCCTGCTGATCGTGAACAACGCCACCAACGTGGTCATCAAAGTGTGCGAGTTCCAGTTCTGCAACGACCCCTTCCTGGGCGTCTACTACCACAAGAACAACAAGAGCTGGATGGAAAGCGAGTTCCGGGTGTACAGCAGCGCCAACAACTGCACCTTCGAGTACGTGTCCCAGCCTTTCCTGATGGACCTGGAAGGCAAGCAGGGCAACTTCAAGAACCTGCGCGAGTTCGTGTTTAAGAACATCGACGGCTACTTCAAGATCTACAGCAAGCACACCCCTATCAACCTCGTGCGGGATCTGCCTCAGGGCTTCTCTGCTCTGGAACCCCTGGTGGATCTGCCCATCGGCATCAACATCACCCGGTTTCAGACACTGCTGGCCCTGCACAGAAGCTACCTGACACCTGGCGATAGCAGCAGCGGATGGACAGCTGGTGCCGCCGCTTACTATGTGGGCTACCTGCAGCCTAGAACCTTCCTGCTGAAGTACAACGAGAACGGCACCATCACCGACGCCGTGGATTGTGCTCTGGATCCTCTGAGCGAGACAAAGTGCACCCTGAAGTCCTTCACCGTGGAAAAGGGCATCTACCAGACCAGCAACTTCCGGGTGCAGCCCACCGAATCCATCGTGCGGTTCCCCAATATCACCAATCTGTGCCCCTTCGGCGAGGTGTTCAATGCCACCAGATTCGCCTCTGTGTACGCCTGGAACCGGAAGCGGATCAGCAATTGCGTGGCCGACTACTCCGTGCTGTACAACTCCGCCAGCTTCAGCACCTTCAAGTGCTACGGCGTGTCCCCTACCAAGCTGAACGACCTGTGCTTCACAAACGTGTACGCCGACAGCTTCGTGATCCGGGGAGATGAAGTGCGGCAGATTGCCCCTGGACAGACAGGCAAGATCGCCGACTACAACTACAAGCTGCCCGACGACTTCACCGGCTGTGTGATTGCCTGGAACAGCAACAACCTGGACTCCAAAGTCGGCGGCAACTACAATTACCTGTACCGGCTGTTCCGGAAGTCCAATCTGAAGCCCTTCGAGCGGGACATCTCCACCGAGATCTATCAGGCCGGCAGCACCCCTTGTAACGGCGTGGAAGGCTTCAACTGCTACTTCCCACTGCAGTCCTACGGCTTTCAGCCCACAAATGGCGTGGGCTATCAGCCCTACAGAGTGGTGGTGCTGAGCTTCGAACTGCTGCATGCCCCTGCCACAGTGTGCGGCCCTAAGAAAAGCACCAATCTCGTGAAGAACAAATGCGTGAACTTCAACTTCAACGGCCTGACCGGCACCGGCGTGCTGACAGAGAGCAACAAGAAGTTCCTGCCATTCCAGCAGTTTGGCCGGGATATCGCCGATACCACAGACGCCGTTAGAGATCCCCAGACACTGGAAATCCTGGACATCACCCCTTGCAGCTTCGGCGGAGTGTCTGTGATCACCCCTGGCACCAACACCAGCAATCAGGTGGCAGTGCTGTACCAGGACGTGAACTGTACCGAAGTGCCCGTGGCCATTCACGCCGATCAGCTGACACCTACATGGCGGGTGTACTCCACCGGCAGCAATGTGTTTCAGACCAGAGCCGGCTGTCTGATCGGAGCCGAGCACGTGAACAATAGCTACGAGTGCGACATCCCCATCGGCGCTGGAATCTGCGCCAGCTACCAGACACAGACAAACAGCCCTCGGAGAGCCAGAAGCGTGGCCAGCCAGAGCATCATTGCCTACACAATGTCTCTGGGCGCCGAGAACAGCGTGGCCTACTCCAACAACTCTATCGCTATCCCCACCAACTTCACCATCAGCGTGACCACAGAGATCCTGCCTGTGTCCATGACCAAGACCAGCGTGGACTGCACCATGTACATCTGCGGCGATTCCACCGAGTGCTCCAACCTGCTGCTGCAGTACGGCAGCTTCTGCACCCAGCTGAATAGAGCCCTGACAGGGATCGCCGTGGAACAGGACAAGAACACCCAAGAGGTGTTCGCCCAAGTGAAGCAGATCTACAAGACCCCTCCTATCAAGGACTTCGGCGGCTTCAATTTCAGCCAGATTCTGCCCGATCCTAGCAAGCCCAGCAAGCGGAGCTTCATCGAGGACCTGCTGTTCAACAAAGTGACACTGGCCGACGCCGGCTTCATCAAGCAGTATGGCGATTGTCTGGGCGACATTGCCGCCAGGGATCTGATTTGCGCCCAGAAGTTTAACGGACTGACAGTGCTGCCTCCTCTGCTGACCGATGAGATGATCGCCCAGTACACATCTGCCCTGCTGGCCGGCACAATCACAAGCGGCTGGACATTTGGAGCAGGCGCCGCTCTGCAGATCCCCTTTGCTATGCAGATGGCCTACCGGTTCAACGGCATCGGAGTGACCCAGAATGTGCTGTACGAGAACCAGAAGCTGATCGCCAACCAGTTCAACAGCGCCATCGGCAAGATCCAGGACAGCCTGAGCAGCACAGCAAGCGCCCTGGGAAAGCTGCAGGACGTGGTCAACCAGAATGCCCAGGCACTGAACACCCTGGTCAAGCAGCTGTCCTCCAACTTCGGCGCCATCAGCTCTGTGCTGAACGATATCCTGAGCAGACTGGACCCTCCTGAGGCCGAGGTGCAGATCGACAGACTGATCACAGGCAGACTGCAGAGCCTCCAGACATACGTGACCCAGCAGCTGATCAGAGCCGCCGAGATTAGAGCCTCTGCCAATCTGGCCGCCACCAAGATGTCTGAGTGTGTGCTGGGCCAGAGCAAGAGAGTGGACTTTTGCGGCAAGGGCTACCACCTGATGAGCTTCCCTCAGTCTGCCCCTCACGGCGTGGTGTTTCTGCACGTGACATATGTGCCCGCTCAAGAGAAGAATTTCACCACCGCTCCAGCCATCTGCCACGACGGCAAAGCCCACTTTCCTAGAGAAGGCGTGTTCGTGTCCAACGGCACCCATTGGTTCGTGACACAGCGGAACTTCTACGAGCCCCAGATCATCACCACCGACAACACCTTCGTGTCTGGCAACTGCGACGTCGTGATCGGCATTGTGAACAATACCGTGTACGACCCTCTGCAGCCCGAGCTGGACAGCTTCAAAGAGGAACTGGACAAGTACTTTAAGAACCACACAAGCCCCGACGTGGACCTGGGCGATATCAGCGGAATCAATGCCAGCGTCGTGAACATCCAGAAAGAGATCGACCGGCTGAACGAGGTGGCCAAGAATCTGAACGAGAGCCTGATCGACCTGCAAGAACTGGGGAAGTACGAGCAGTACATCAAGTGGCCCTGGTACATCTGGCTGGGCTTTATCGCCGGACTGATTGCCATCGTGATGGTCACAATCATGCTGTGTTGCATGACCAGCTGCTGTAGCTGCCTGAAGGGCTGTTGTAGCTGTGGCAGCTGCTGCAAGTTCGACGAGGACGATTCTGAGCCCGTGCTGAAGGGCGTGAAACTGCACTACACATGATGACTCGAGCTGGTACTGCATGCACGCAATGCTAGCTGCCCCTTTCCCGTCCTGGGTACCCCGAGTCTCCCCCGACCTCGGGTCCCAGGTATGCTCCCACCTCCACCTGCCCCACTCACCACCTCTGCTAGTTCCAGACACCTCCCAAGCACGCAGCAATGCAGCTCAAAACGCTTAGCCTAGCCACACCCC
>k119_186801- ColE1- T7
CGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTCAGGGGGGCGGAGCCTATGGAAAAACGCCAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTTGCTGGCCTTTTGCTCATTAGGCACCCCAGGCTTTACCCGAACGACCGAGCGCAGCGAGTCAGTGAGCGAGGAAGCGGAGAGCGCCCAATACGCAAGGAAACAGCTATGACCATGTTAATGCAGCTGGCACGACAGGTTTCCCGACTGGAAAGCGGGCAGTGAGCGGAAGGCCCATGAGGCCCAGGAATTCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT
>k119_227827- HBA1 and HBA2 Beta Globin Sequence
CGATATGGTAAAATTTATTCAGACTGTAAAGTAGGACCATAGGGGAATTAAGGAAGGGAGGAAACCATGGAAGGCATCTCTTACATCTGGGCTAATCCGAAAGTAGATTTGATTTATCCCAACGTCAAGGCCGACCTTACCCCAGGCGGCCTTGACGTTGGTCTTGTCGGCAGGAGACAGCACCATGGTGGGTTCTCTCTGAGTCTGTGGGGACCAGAAGAGTGCCGGGCCGCGAGCGCGCCAGGGTTTATGCTTGGGGCGCGGGGGCACGCCCGGCCGGGCGGCGCTCATTGGCTGGCGCGGAGCCCGGGGCGCGGCCTGGACCGCAGGGGAGTCCCGGGCGGGGCCTGCGCGGGGCCGGGCGGCGGGCTCGGCCTCCCCTGGGGGTGCACGCGGGGCGGGGGCCAGGACGTCTCCACCCTCCACCCGCCACTCCACTCCCGCCCATCCCGCTCGCCCGGGAGCAGGGCGGTAGCGGCGCTGGCGGGGCCGGCCCGTTGGGGTCGGGCGCTGTCGGCTCGTGCACCCCGGAGCGCAGCGCCACCCTTTCCTTTCGGGCGCCGGAGCGTCTCTAGCGAGGGAGAAAGTCAGCCCGCACCCCCGCCCCGGCCTGGCACGCGCTGGACGCGCATCGACTCCAGCGGGATCGGGGAACACACGGGCGAGCGAGTGCGAGCCGGGAGGCTTCGCCCAATCCTGGGGCGGAGAGGAATGCGCGCACCCGGACGCCCTGGCCCATAGGAACTCAAAAGAATTTCTGCGCAGAGCCCCTCCTGCTCTCCAGCCTCGCTTCCTGCAGCTCCCTTTCCCTCTGGCGATAGTCACTAGT
>k119_230647 SV40 poly A signal and F1 Origin of Replication
AAAAAAAAAAGAAGAGCTCCAACCGGTGTGGTAGCTCCGCCGTTTAACATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGAGATCCAATTTTTAAGTGTATAATGTGTTAAACTACTGATTCTAATTGTTTGTGTATTTTAGATTCACAGTCCCAAGGCTCATTTCAGGCCCCTCAGTCCTCACAGTCTGTTCATGATCATAATCAGCCATACCACATTTGTAGAGGTTTTACTTGCTTTAAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCAATTGTTGTTGTTAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTAACGCGTAAATTGTAAGCGTTAATATTTTGTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGACCG
>k119_25735 Neo/KAN HSV TK
CTTGGGGGGTGGGGTGGGGAAAAGGAAGAAACGCGGGCGTATTGGCCCCAATGGGGTCTCGGTGGGGTATCGACAGAGTGCCAGCCCTGGGACCGAACCCCGCGTTTATGAACAAACGACCCAACACCGTGCGTTTTATTCTGTCTTTTTATTGCCGTCATAGCGCGGGTTCCTTCCGGTATTGTCTCCTTCCGTGTTTCAGTTAGCCTCCCCCTAGGGTGGGCGAAGAACTCCAGCATGAGATCCCCGCGCTGGAGGATCATCCAGCCGGCGTCCCGGAAAACGATTCCGAAGCCCAACCTTTCATAGAAGGCGGCGGTGGAATCGAAATCTCGTGATGGCAGGTTGGGCGTCGCTTGGTCGGTCATTTCGAACCCCAGAGTCCCGCTCAGAAGAACTCGTCAAGAAGGCGATAGAAGGCGATGCGCTGCGAATCGGGAGCGGCGATACCGTAAAGCACGAGGAAGCGGTCAGCCCATTCGCCGCCAAGTTCTTCAGCAATATCACGGGTAGCCAACGCTATGTCCTGATAGCGGTCCGCCACACCCAGCCGGCCACAGTCGATGAATCCAGAAAAGCGGCCATTTTCCACCATGATATTCGGCAAGCAGGCATCGCCATGGGTCACGACGAGATCCTCGCCGTCGGGCATGCTCGCCTTGAGCCTGGCGAACAGTTCGGCTGGCGCGAGCCCCT
The assemblies are broken where the Pfizer plasmid begins to share homology with the library construction plasmids but is contiguous for all of spike and other non-polluted regions of the Pfizer plasmid.
Another paper was forwarded to me by Jonathan Gilthorpe from Odak et al. This is also cited in the Chakraborty et al paper as the ‘German Study’.
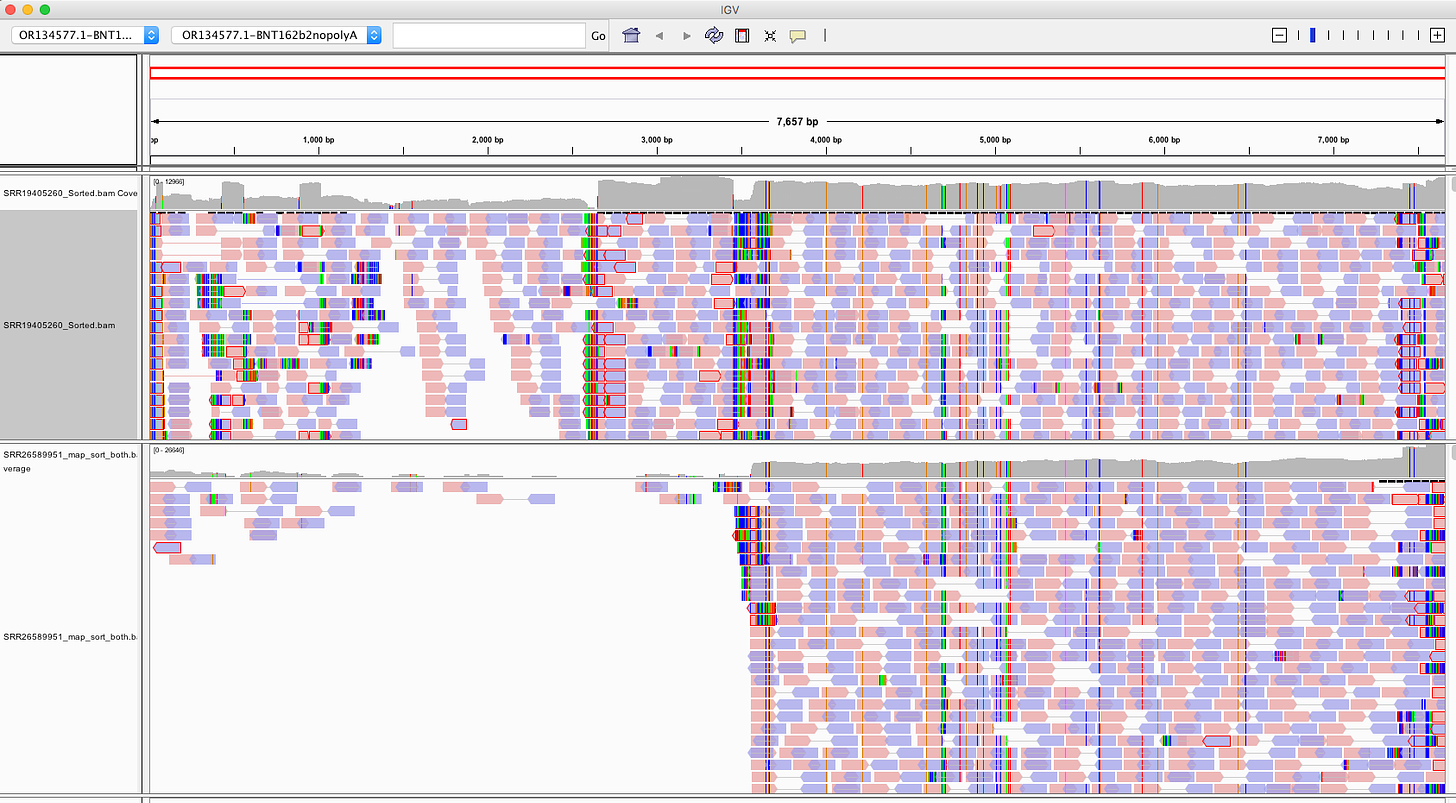
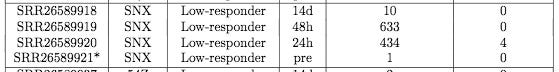
Below (Bottom track) is SRA accession number SRR26589951. Top sample is one of the most contaminated samples from Ryan et al.
Many other 24hour, 48hr and 14 day samplings exist. If SRAtools is giving you headaches, you can look these up on the ENA link above and they are much easier to download.
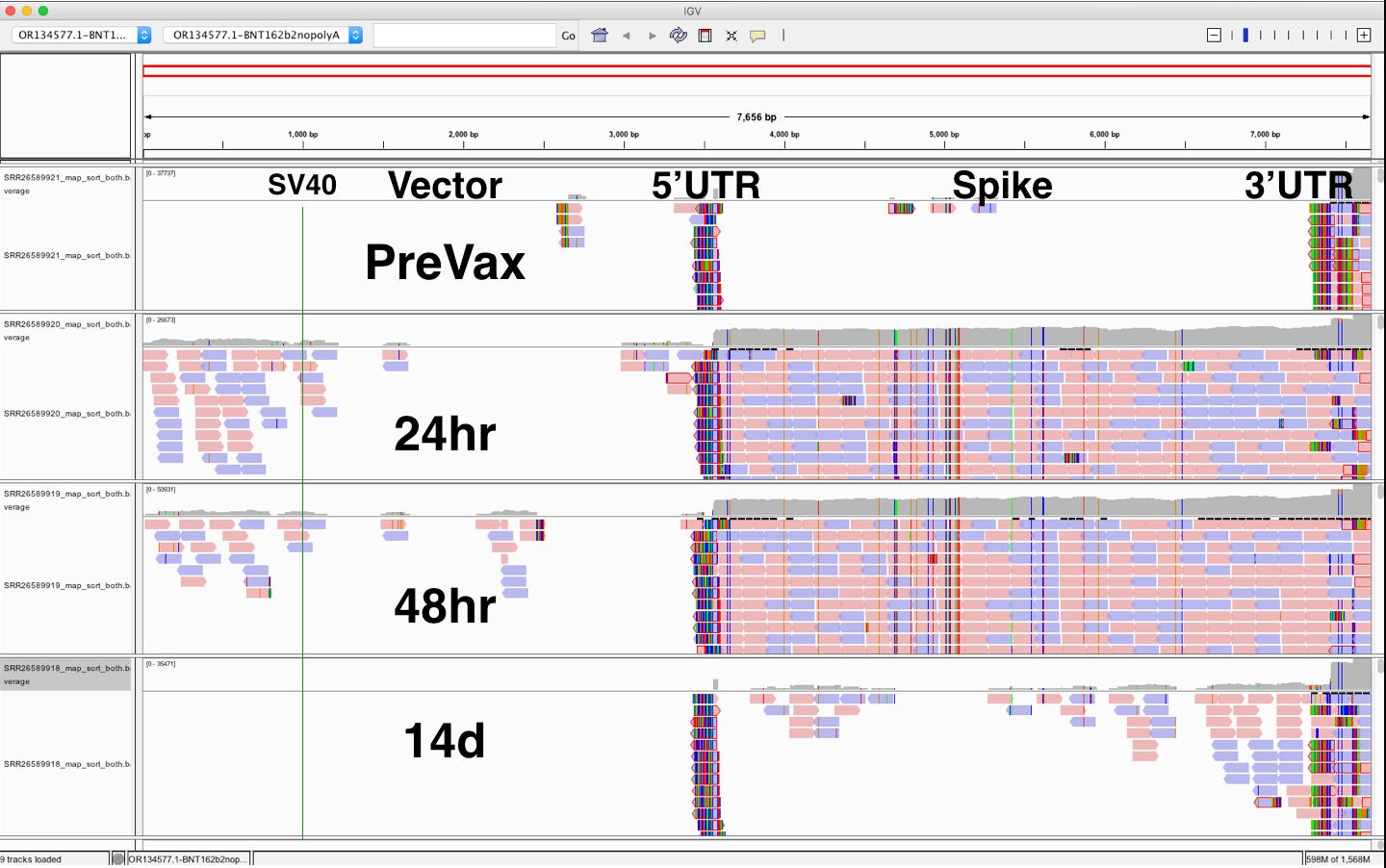
If we take these 4 samples from the ‘German study’ in Chakraborty et al and load them into IGV what do we see?
You can see the 5’UTR and the 3’UTR which are human beta globin 5’UTR and Human Mito 3’UTR both recruit human reads into the mappings. The SV40 is present in Day 1 and Day 2.
These reads were trimmed with quality restrictions at -q 20 and -m 30
cutadapt -a AAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA -A AAGTCGGATCGTAGCCATGTCGTTCTGTGAGCCAAGGAGTTG -q 20 -m 30 -o TRIMMED/SRR26589919_1.trim.fastq.gz -p TRIMMED/SRR26589919_2.trim.fastq.gz SRR26589919_1.fastq.gz SRR26589919_2.fastq.gz
Only mapped reads were output through Samtools.
bwa mem -t 96 OR134577.1-BNT162b2_no-polyA TRIMMED/SRR26589919_1.trim.fastq.gz TRIMMED/SRR26589919_2.trim.fastq.gz | samtools view -@24 -b -F 4 | samtools sort -@ 24 -o SRR26589919_map_sort_both.bam
What are DNA depletion steps? Usually DNaseI or Actinomycin D. Since I asked the authors about DNaseI and they didnt reply with a confirmation but claiming a DNA depletion step was used, Im going to assume they are using Actinomycin D as most stranded RNA-Seq kits take this approach as they don’t want residual DNaseI around during cDNA synthesis.
Conclusions
Both studies show signs of DNA contamination in the blood of vaccinated patients. Their exact quantity and duration is obscured by DNA depletion steps in the RNA purification process and background plasmids in the Ryan et al study. The Odak study is clean of these background plasmids. The watson strands exist higher than the expected background rate in the BNT162b2 plasmid vector (1000-2600bp, 560-900bp, 87-420bp) demonstrating this is DNA not RNA. SV40 promoter sequences are detected. Considering these contaminants are wrapped in LNPs and are still detectable 48 hours out using methods designed to eliminate them should be urgent grounds for immediate investigations into how long this DNA persists in absence of DNA depletion protocols. The purification tools used to capture this DNA need to be heavily scrutinized as Ethanol Precipitations and many DNA purification kits will eliminate small DNA or further digest them with DNaseI.















Why is this stuff still being injected into unwitting human beings? Are the regulators completely incompetent, or are they in Satan's grasp.
We have checked some cv19 ab levels. All our cases are gravely x50-250 (of positive immunity treshold) hyperimmune for spike abs having NONE env or nucleocapsid abs. These mRNA vaxxed cases mostly have not had vxx or cv19 disease for last year. This must indicate vxx SPIKE PERSISTENCE, probably continuous production of spike, indicating inability of getting rid of n1-meth-pseudoU mRNA, or integration event in some tissues or gut bacteria, me wondering.
...btw..Measuring abs is strongly discouraged and labs here mostly report as +/- not giving real values even when a clinician asks.
Our national health authorities must be aware of this but still pushing it for already hyperimmune and misinformed ppl.
My message is that our few ab measurements support integration theory.