A few folks asked if Pfizer or Moderna could use their IP portfolios to arrest testing for the vaccine contamination in patients.
It’s a good question. I will attempt to answer it by highlighting the patents that are likely the most relevant and if anyone finds other patents they think are relevant, please put them in the comments below.
The first 3 patents to look at are the patents that Moderna and Pfizer are in legal battles with each other over. These are known in the legal space as the ‘127, ‘574, and ‘600 patent.
For patents you need to look over the independent claims. When these are multipart claims, you only need to not perform one of the parts to NOT infringe the patent.
Let’s look over the claims in the legal battle-
10,702,600 (the “’600 patent”),
qPCR testing a patient will not involve formulating anything in a lipid nanoparticle. Were clean of this.
10,933,127 (the “’127 patent”)
qPCR testing will not involve administering anything to a subject.
We are clean of this.
qPCR testing will not be producing a polypeptide of interest in a cell in a subject.
So the main patents that are in dispute between Moderna and Pfizer have no claims that prevent qPCR of patients.
OK- So are there any claims to their sequence of the spike sequence.
If you search NCBI for BNT162b2, 3 Sequences stand out.
The first is the sequence we submitted to GenBank. This is the sequence of their plasmid.
The second and third speak to the below patent application. It is not issued yet.
https://patents.google.com/patent/WO2021214204A1/en?oq=WO2021214204
https://s3.documentcloud.org/documents/22266025/moderna-v-pfizer.pdf
This speaks to a medical preparation with certain sequence IDs. qPCR will not generate something with a 5’ Cap.
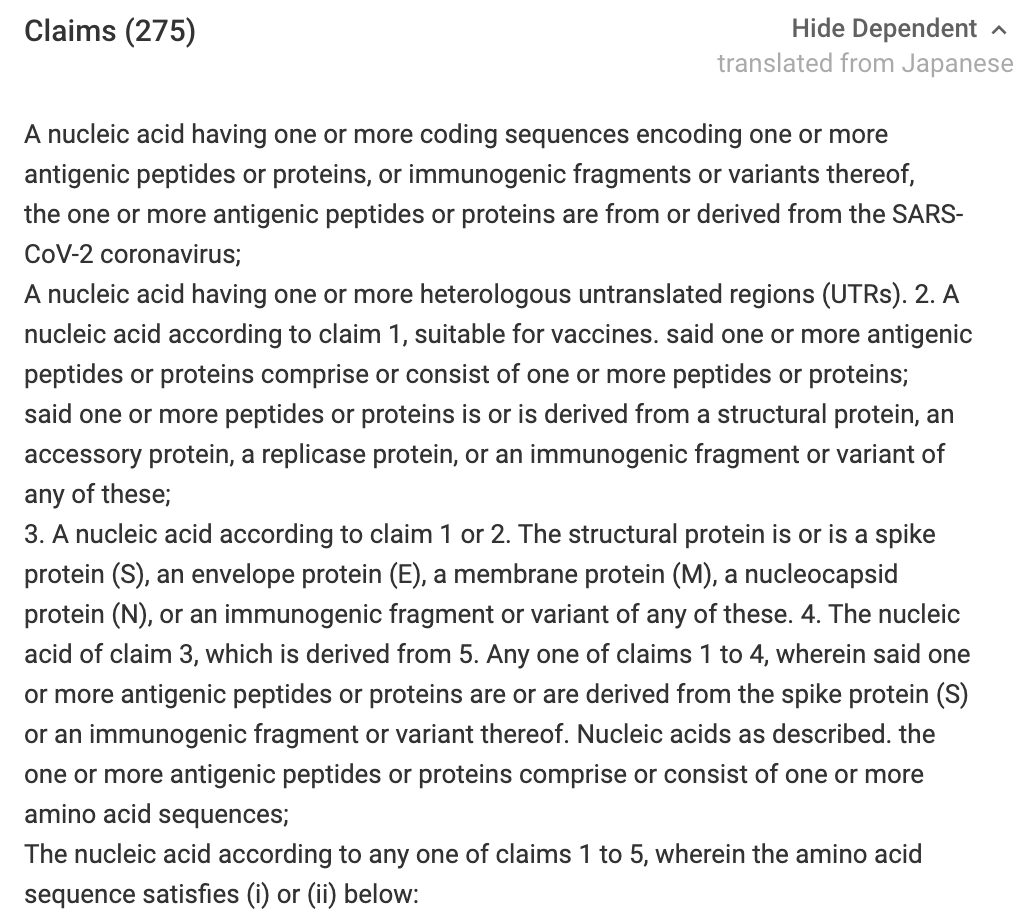
Also be sure to search NCBI’s patent database with the sequence itself. You’ll find this Japanese patent.
Notice the claims speak to encoding language and the DREAM PCR paper addresses these types of claims head on. It also requires the nucleic acid have UTRs which none of our PCR products will have.
So far, nothing that prevents qPCR of patients but lets say they amend claims or we missed a particular composition of matter related to the Spike protein and something submarines later.
We have been here before.
This is not our first Gene Patent Rodeo. We ran a clinical sequencing company that had to navigate 1000s of gene patents just to simply sequence a sick kids genome.
We came up with this method to navigate them and were never challenged despite 5 years commercially sequencing in the field.
It surprisingly was published by Nature Biotechnology and is likely the only time Murray Rothbard and Hayek are cited in Nat Bio.
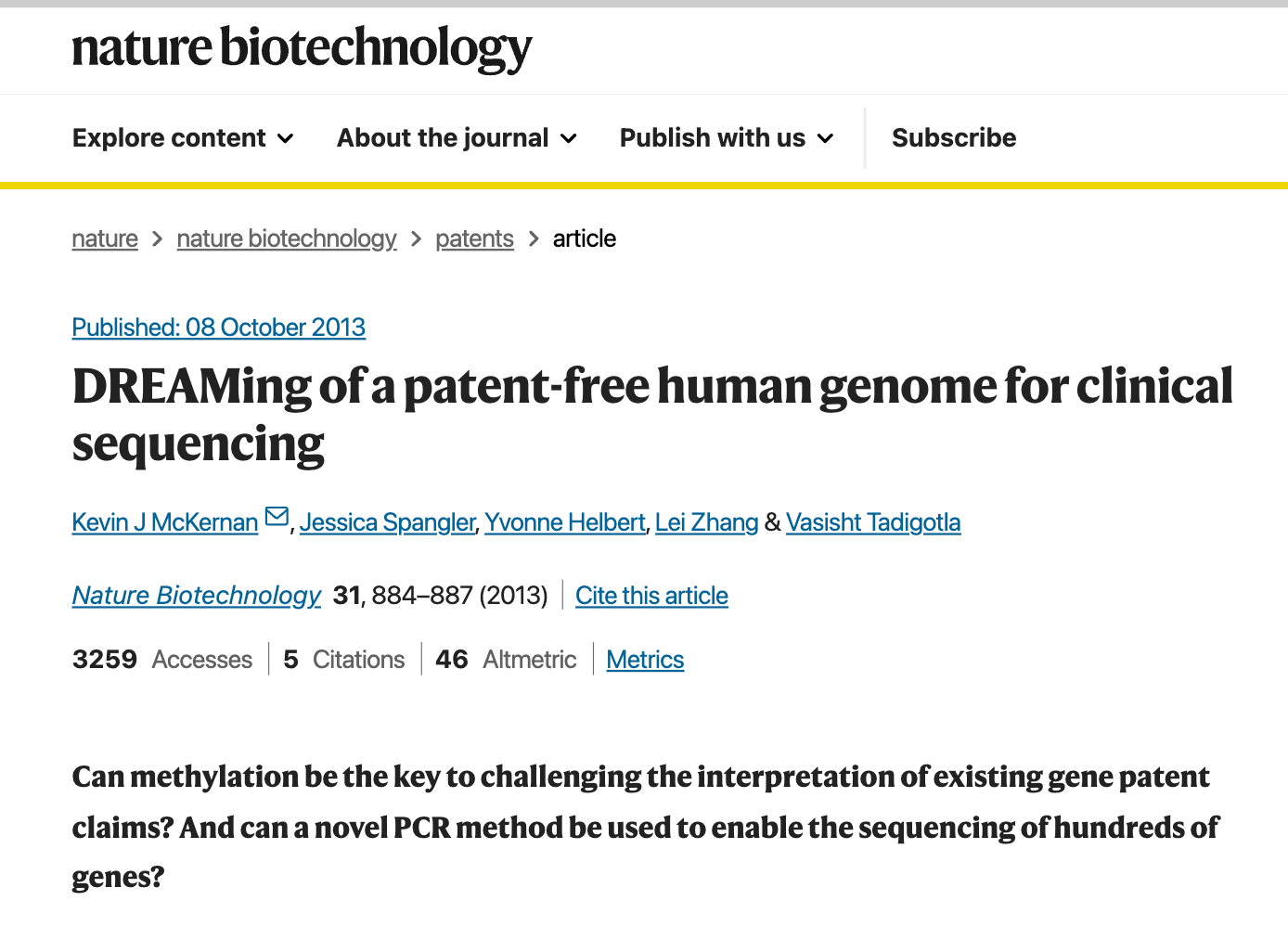
The qPCR kits we designed to target the vaccines, utilize this technology. In affect, when we amplify a target sequence, we replace Cytosine with MethylCytosine (much like the vaccines swap out Uracil with N1-methyl-Pseudouracil. In some applications we use HydroxyMethylCytosine.
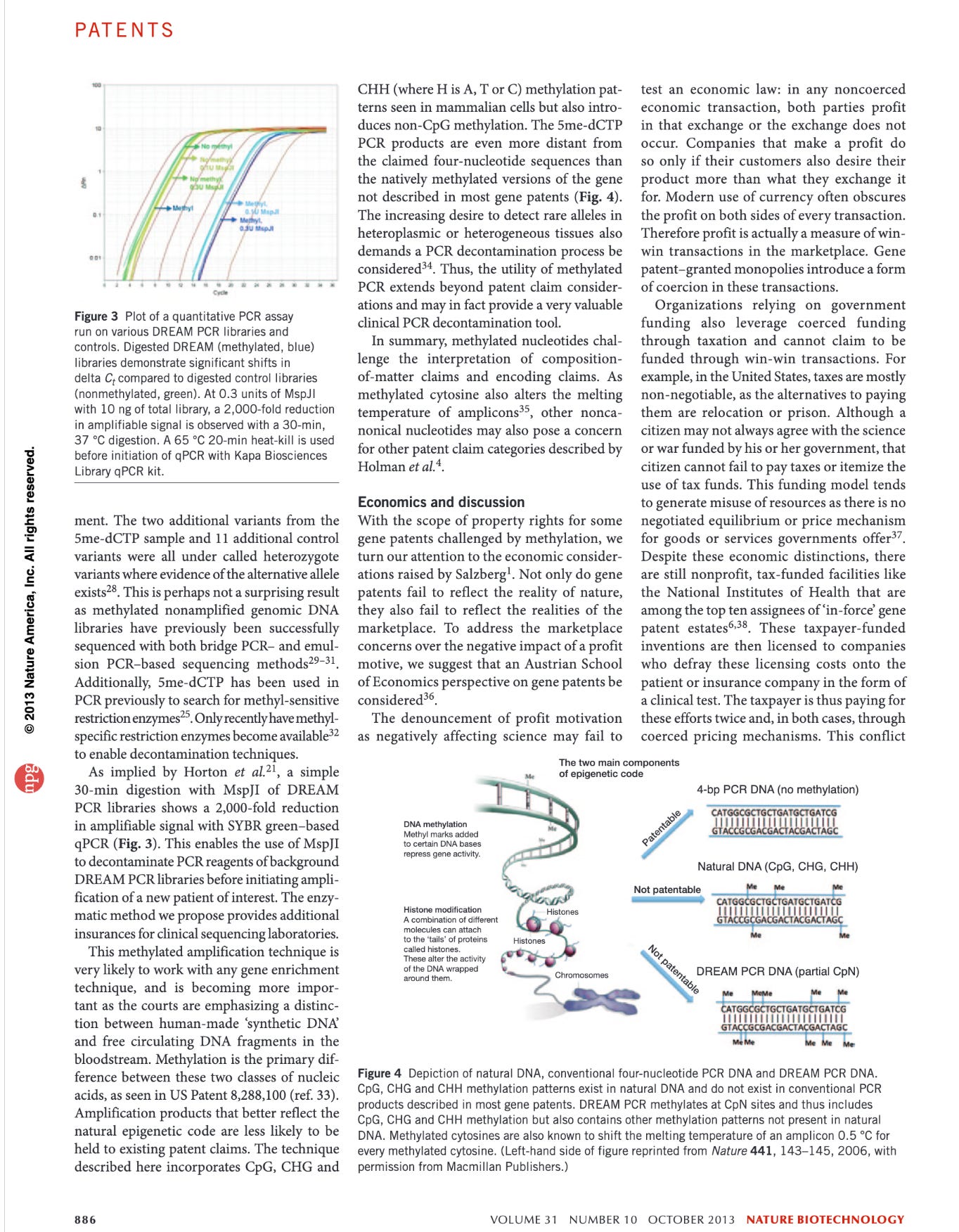
As a result the DNA we amplify is not the same as the DNA claimed in most Gene Patents. It is materially different.
We took this approach one step further and published a tweak on this in PLOS One.
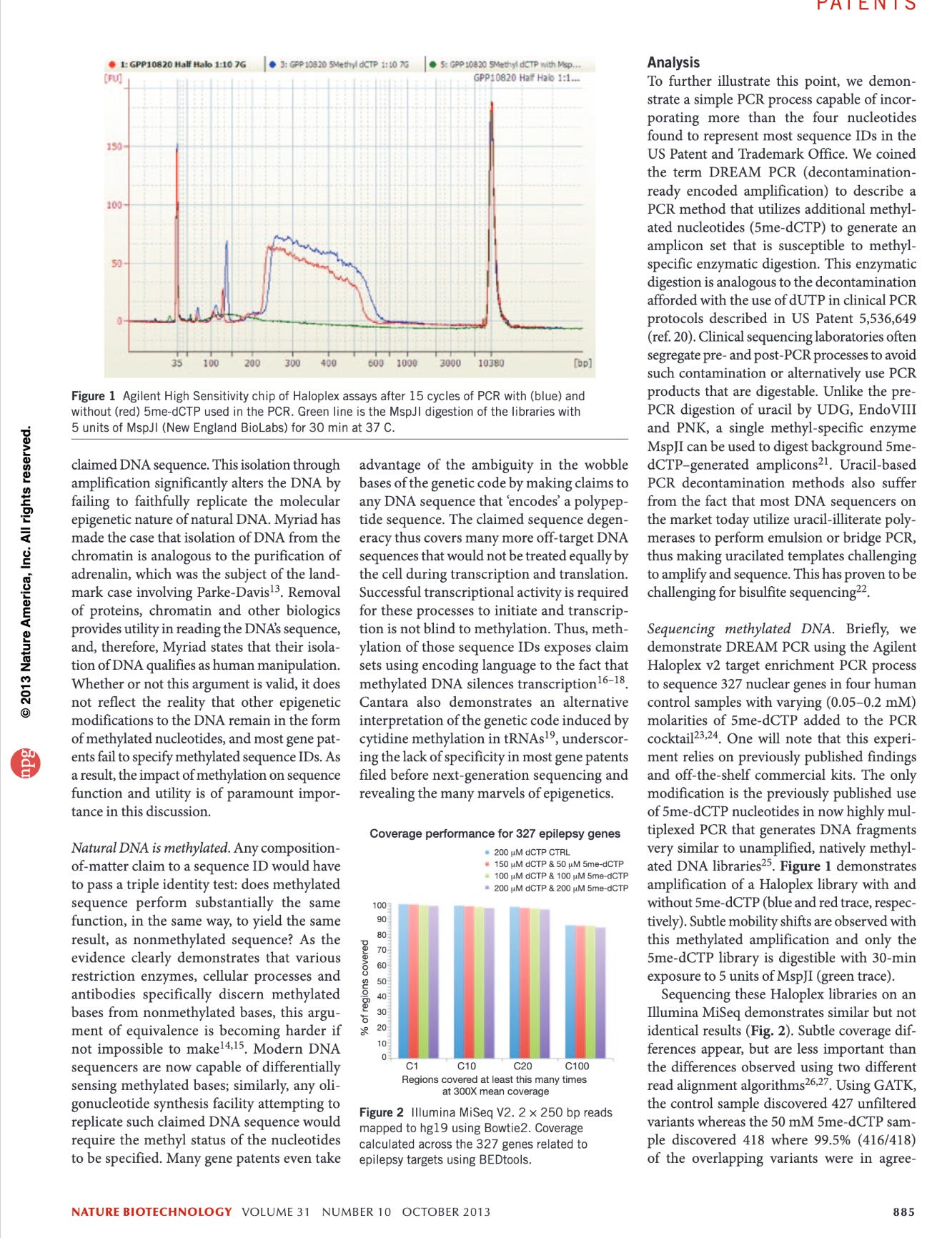
These papers are actually relevant as adding methyl groups to bases in DNA radically alters the melting Temperature of the DNA. This is an important point that can be leveraged in PCR. When the gDNA is relative lightly methylated, after your 1st strand synthesis in PCR, your templates now all have a higher melting temperature. As a result you can raise the annealing temperature in PCR to favor more stringent replication of you target.
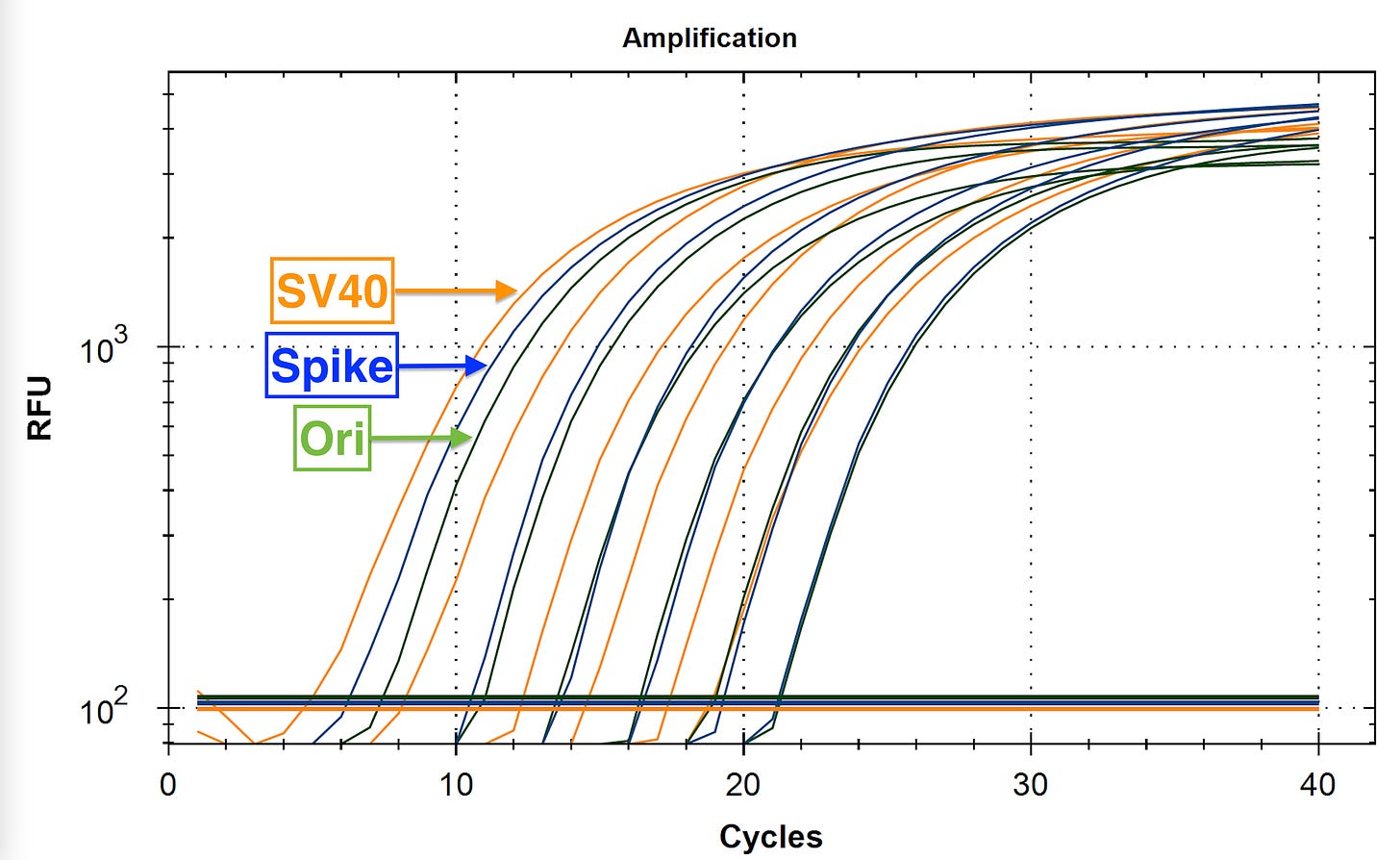
A few of our detractors on Twitter were not very well read on the PCR art and were trying to make the claim that our SV40 primers didn’t work (Despite the CT scores shown in David Speichers manuscript).
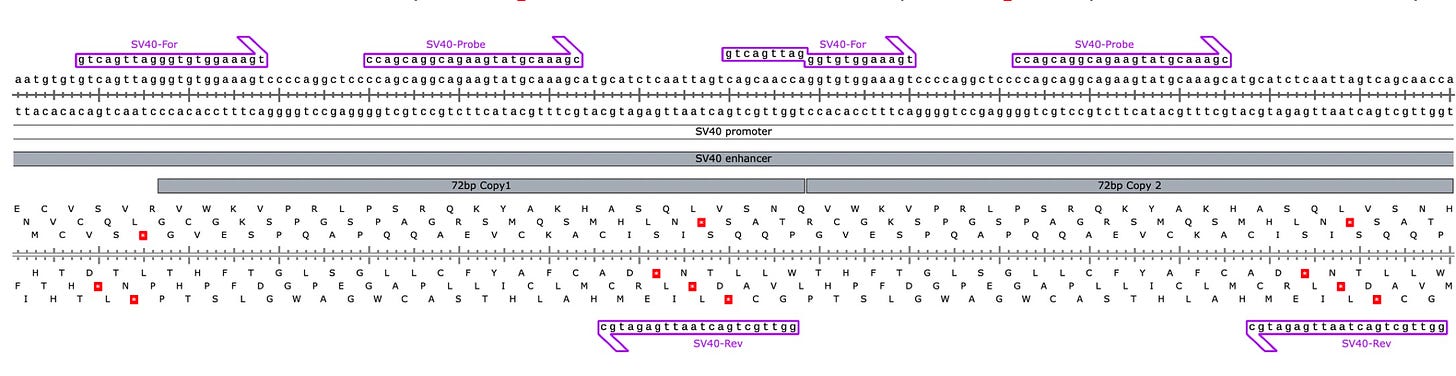
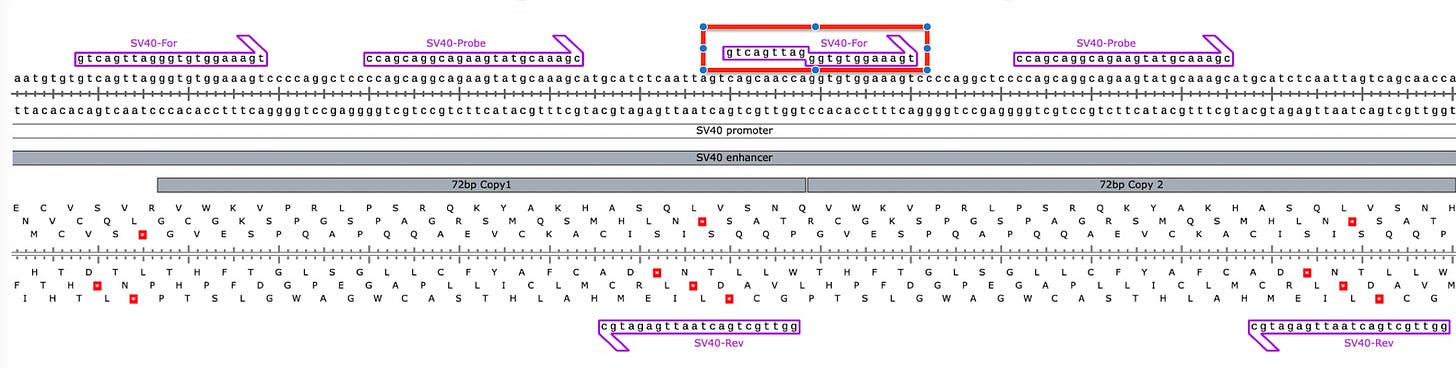
The came to this in silico conclusion because one of the SV40 Forward primers sits on the junction of the tandem 72bp repeat in the SV40 Enhancer. Its important to note that this same primer exists all the way on the left side of this image such that the internal forward primer isnt actually needed but if we can get it to contribute then we will have 2 active PCR amplicons contributing to signal PCR.
Since the SV40 Enhancer is the most likely region to integrate, we dont want to leave any signal on the table. As a result an amplification strategy that can amplify both copies of the 72bp Enhancer will have twice the sensitive as an assay that just targets one.
So the detractors zero’d in on the Forward Primer in the middle (Red box) that has a Tail that doesn’t perfectly match the template. It has a 12 bp homology on the 3’ End and a 9bp base pair tail that isn’t complimentary until the 1st cycle of PCR is complete.
Below in red is the primer they have criticized.
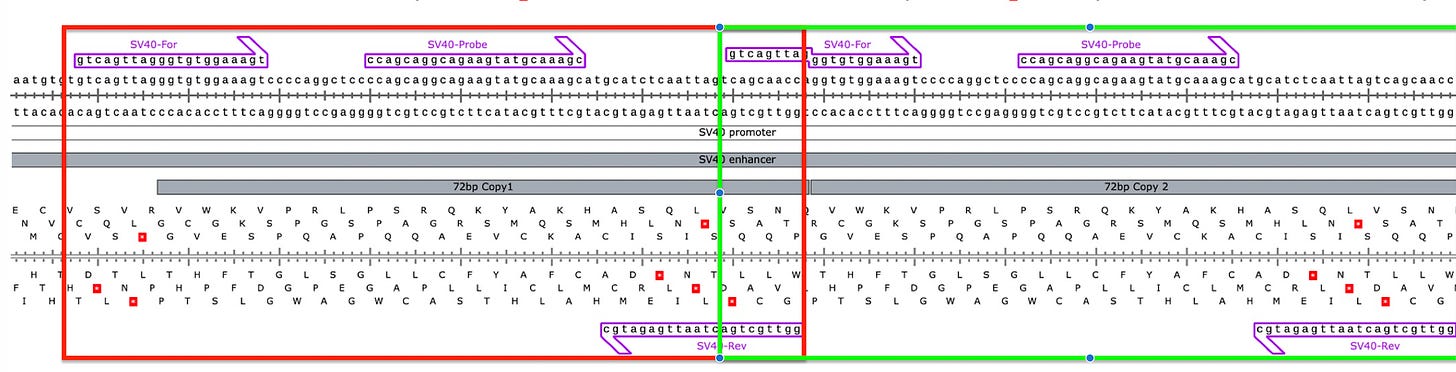
What actually happens in qPCR is that this design breaks open the dual 72bp Enhancer into 2 independently amplifiable targets (Red and Green) that are each probed to provide 2X the signal.
Normally that SV40 Forward 12mer will only have a Tm of 48-50C. But you will notice the 3’ end of that forward primer has 6 Gs in it. Once the reverse strand is made with 6 methyl Cs’ the Tm Jumps to 59C. So the 1st cycle is high stringency and low efficiency but the second cycle takes off.
This is something known as Tailed PCR. After the 1st cycle of PCR, The Tails become incorporated as the reverse strand reads over your top strand primer. If in the process of generating your reverse strand you are decorating the template with stickier bases (higher Tm) then you can make shorter primers participate with higher efficiency than you had on the first strand synthesis.
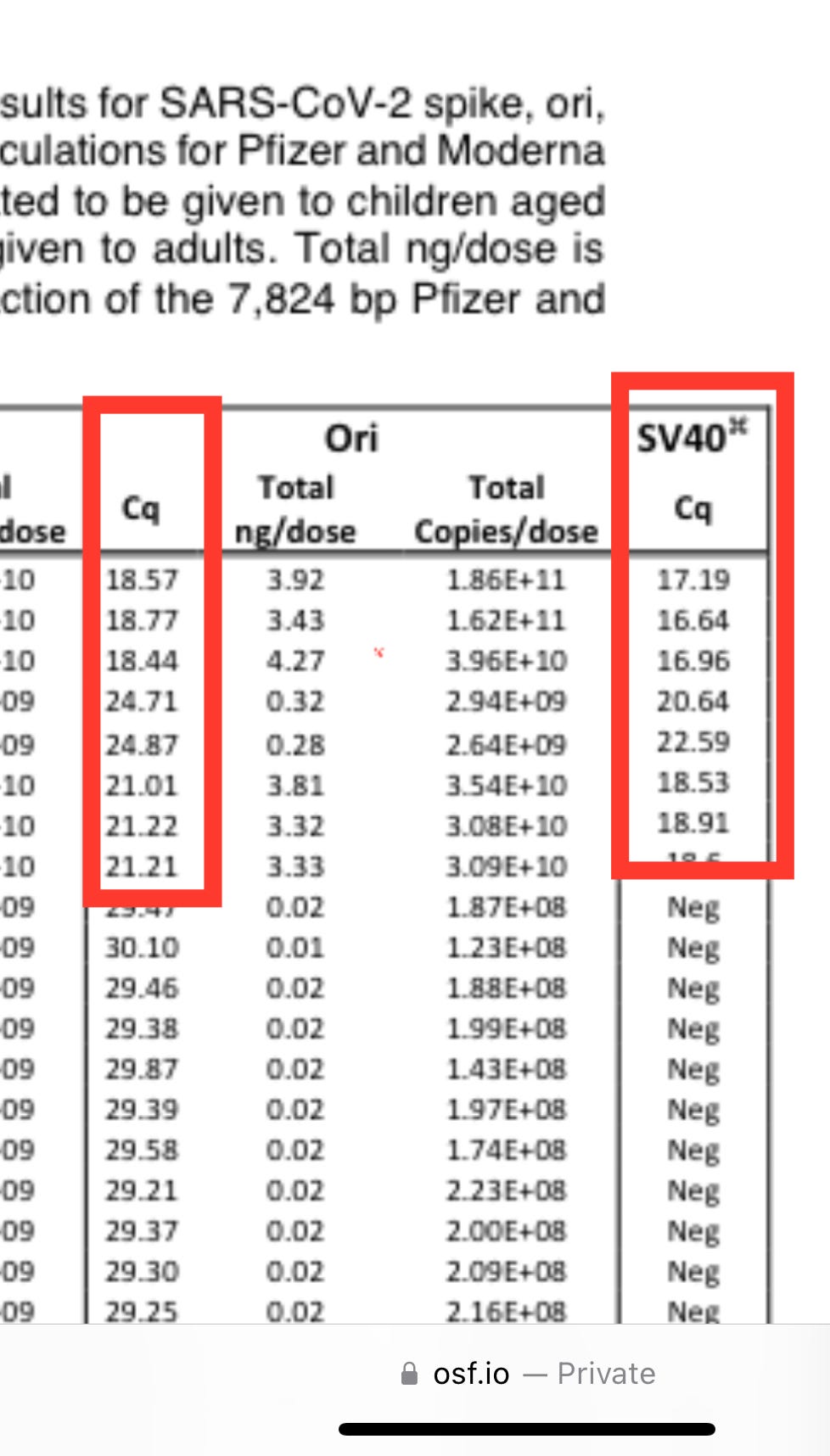
This is why the SV40 amplification has lower CT scores (more signal) in
paper.This wasn’t good enough for them so they moved on to pointing out that we didn’t have a standard curve for SV40 in the paper. This is true as we didn’t prioritize it. Moderna doesn’t have SV40 so it wouldn’t have added much to the paper to delay it and wait for that Standard to be synthesized. It only lights up on the Pfizer vaccines so the paper was best focused on the Ori and Spike amplicons.
Why didn’t we have a standard curve for SV40? Was it some nefarious conceal?
Nope.
IDT had a hard time synthesizing a 72bp tandem repeat. In fact they refused to do it.
So we had to tweak a few things to make this work at IDT and now have the gBlock and have run standard curves.
If you run a serial dilution on the Pfizer vaccines below, you can see the SV40 amplicon is the most sensitive assay we have due to this multiplex targeting of the SV40 tandem repeat.
So while the detractors assumed we were qPCR gimps, what is actually going on here is likely a patentable approach and their tweets are all evidence that this is non-obvious and likely inventive.
We already have patents on DREAM-PCR in multiple jurisdictions so we don’t need their evidence but I thought this was a good story to share.
Back to the concern over Pfizer and Moderna using IP to shut down the clinical testing of victims..
1)The region of Spike we target is the same in Moderna, Pfizer and J&J. Will be hard for anyone of them to claim it.
2)The Ori and SV40 Amplicons are outside of their grasp as these are age old vector sequences.
3)DREAM PCR likely navigates any composition of matter claims, or encoding claims they make on spike and our PCR assays took this into consideration from Day 1.
Not our first Rodeo.














Thank you for your great work!
You also need to learn about the exhaustion doctrine of patent rights and the exclusion doctrine of patent infringement related to medical practices. In the United States, both cases have been decided in the Supreme Court. It must be noted that biotechnology patents are an exception to the medical practices.
There is certainly a risk of submarine patents, where patent trolls who focus on your implementation later acquire a patent right based on your product. In this case, it is important to find a patent including the DNA or RNA base sequence to be a target of the planned PCR. If it is or it has a divisional application or a continuation application, you need to strongly pay attention to them, but if these applications have not been filed, the risk of a submarine patent may become low.
If a sample extracted from patient is to be identified using PCR, patents related to PCR itself should also be very important. A sample extraction method using PCR and a measurement method using PCR are exemplified.
The samples extracted from patient are parts of a vaccine, but they have already been altered somehow in the body, so they probably won't look exactly like the patents disclosed by Pfizer, Moderna etc. If the plasmid DNA fragments are to be extracted, it is highly unlikely that such arbitrary fragments would be granted patent rights. This is because the very idea of patenting plasmid DNA fragments itself is unproductive and ridiculous.
In my opinion, the method of extracting plasmid DNA fragment (including genomic integration site) from the body of an mRNA vaccine recipient and amplifying and detecting it (diagnosing medical conditions) using PCR seems to be patentable in itself (lol). This is because the very idea that plasmid DNA fragments existed in the body was an improbable idea until now.
In any case, it is important to pay attention and search for patents, and it is great to publicly call out their existence.
I express my utmost respect for your work.
These guys must HATE you for this