
This is a running Lab Blog on a series of experiments to assess if HpLVd infection of Cannabis can satisfy Koch’s Postulates
To recap-
Collin Palmer’s UK Cheese plant tested positive in the leaves for HpLVd during vegetative growth back in September, 2023. After flowering, this plant turned deep purple (anthocyanin production) and then proceeded to test negative in the canopy ~4 weeks later on purple leaf tissue.
Why do we care about this anthocyanin production? Xu et al performed transcriptome analysis with Hop Stunt Viroid infection in sweet cherry and found genes involved in anthocyanin production up-regulated. This is not the same viroid as HpLVd but it may be predictive of plant responses with other related viroids.
UK Cheese being HpLVd positive in the leaves during vegetative growth and then turning HpLVd negative in the anthocyanin rich leaves during flowering is a curious finding. This led us to get access to the roots to see if the plant had cleared the viroid entirely or merely constrained HpLVd to the roots as we have seen in the past with Jamaican Lion cultivars?
The Roots were in fact HpLVd positive and the green tissue in the canopy is also positive but at higher HpLVd CT than the roots (higher CT = less HpLVd). Purple tissue continues to test HpLVd negative (>CT35).
This was first observed with Jamaican Lion (JL) last year. I presented at CannMed on a large multi-tissue RNA-Seq study we performed on Jamaican Lion through the course of an HpLVd infection. This cultivar was never positive in the leaves. After injection of a ground up leaf slurry from an infected vegetative plant, JL only became HpLVd positive in the roots. This mode of infection isn’t a purified viroid and any symptom transmission could not be solely attributed to HpLVd transmission.
To complement the UK Cheese data, we infected Jamaican Lion clones with purified HpLVd circRNA (synthesized). It is important to emphasize that the molecules were not ‘isolated’ from a mixture of other biologicals but were chemically synthesized ground up from scratch with no other biological contaminants. There is no “it has never been isolated” canard with synthetic genomes.
These Jamaican Lion clones became positive in the roots 5 days post infection (dpi) and were negative in the canopy at 5dpi.
We injected 723 billion to 7.23 trillion circRNAs into the stems of 2 JL clones. This large quantity of injection could simply manifest as a dilution effect into the plant given the CT scores in the roots are lower than what was injected. The canopy is HpLVd negative 5dpi suggesting an uneven dilution effect of the injected viroid.
At 5dpi the Jamaican Lion clones were switched to 12 hour lighting (flowering).

After 5 days of flowering (10dpi), HpLVd can now be detected in the leaves, petioles and meristem with RT-qPCR.
So how do we discern simple dilutions of the circRNA from replicative RNA?
First you need to know a little bit about how these viroids replicate in the host. They are too small (256bp) to code for any polymerases so they must rely on host replicative machinery.
HpLVd uses an Asymmetric rolling circle amplification, where the circular RNA template is replicated by a host polymerase into linear concatemers depicted in blue. These are used as templates for RNA polymerases, RNases and Ligases to make further circles which repeat the process. The exact enzymes that perform this replication in Cannabis have not been identified but the genome sequencing of Jamaican Lion has created a list of candidate orthologs to plants where these processes are better understood.
In order to understand the replicative potential of injected synthesized circRNA, monitoring the linear to circular RNA ratio becomes important. The input synthetic circRNA will have a constant circular to linear ratio. Any circRNA that is actively replicating will have a skewed circular to linear ratio.
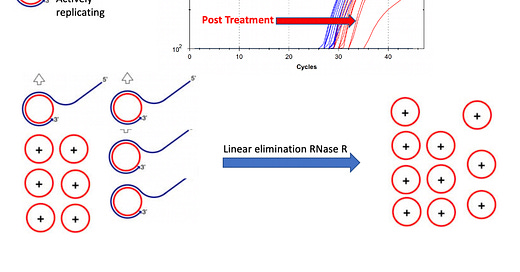
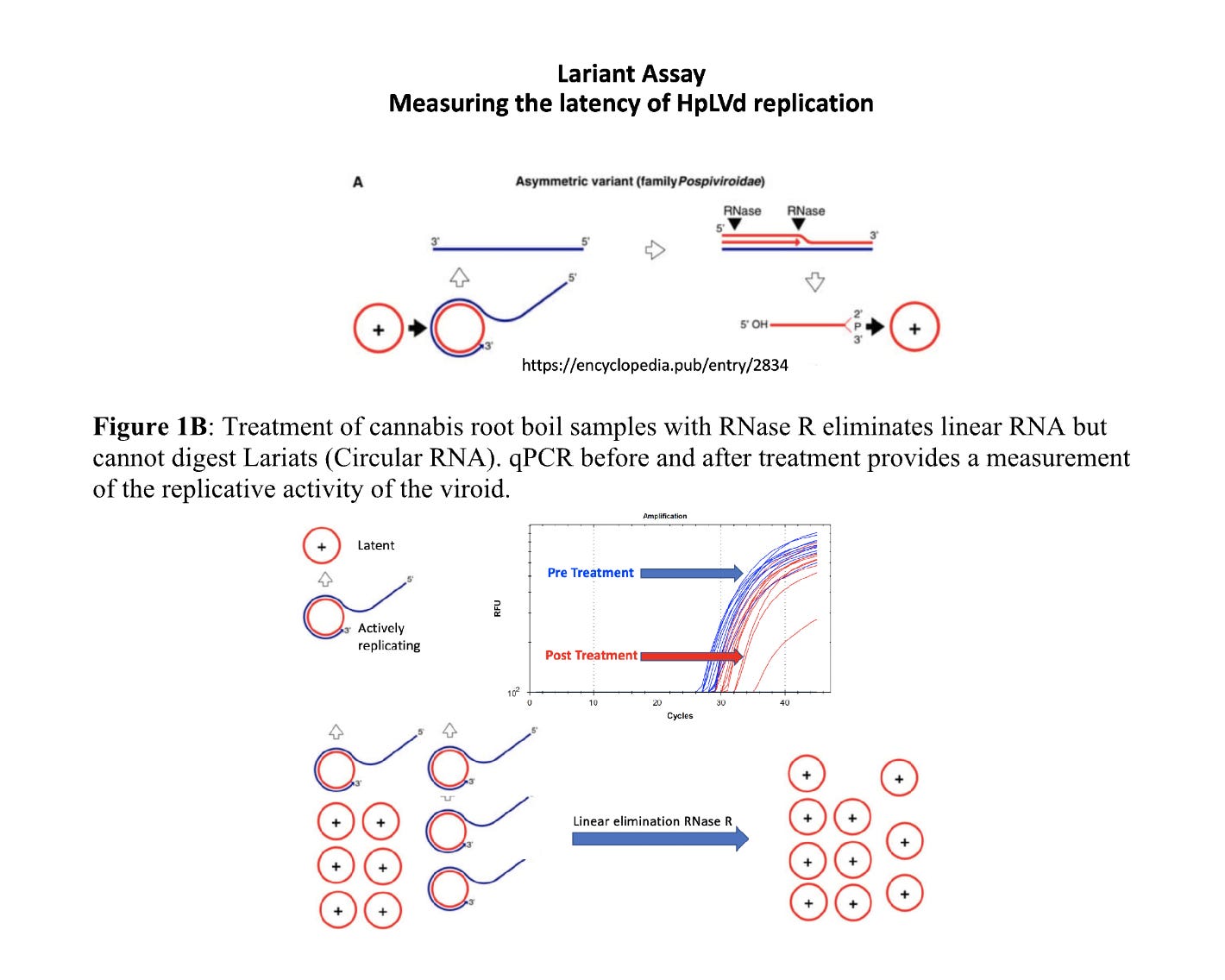
To address this, we developed a Lariat assay to ascertain the replicative stage of the viroid. This leverages RNaseR which only degrades linear RNA and cannot hydrolyze circular RNA.
By running this assay on the injected synthesized circRNA and comparing it to the RNA found in the plant we can ascertain if the injected viroid is replication competent. There should be linear RNA forming after infection that is detectable with RT-qPCR.
First we survey this assay on the biologically infected UK Cheese to understand the linear to circular variation compared to the synthesized control.


The control sample at the bottom of Figure 7 is pure circular RNA. About 5% of this is believed to be linear according to the GeneScript QC. All the samples with larger Cq offsets than the circRNA control have more linear RNA than circular RNA demonstrating replicative HpLVd in the plant. This half CT difference on the circRNA control performed with and without RNaseR is likely within the error of qPCR and pipetting variances. ddqPCR is needed to better quantitate less than 1 CT differences.
UK Cheese is a naturally infected plant and serves as a biological baseline for what we should see in the injected plants if the viroid replicates.
As we turn to the infected JL plants that are 5 days into flowering and 10dpi we see much more linear RNA being synthesized by the plant. The linear to circular ratio is largest in the canopy.

This is an interesting observation. At 5dpi the viroid was only detectable in the roots implying simple dilution through the plant had to concentrate the viroid in one tissue. After 5 days of flowering, we can see the viroid has moved to the canopy and has a much different linear to circular RNA ratio than the roots. The CT values in the canopy are also lower than in the roots implying more HpLVd genome is being generated. This negates an alternative hypothesis of plant RNase activity simply nicking the circular RNA as it moves throughout the plant and thus creating RNA that is RNaseR sensitive.
To assess the sensitivity of the assay, both Limit of Detection analysis (LODs) and Deep RNA sequencing was performed.
8 additional biologically infected plants (different cultivars) that tested positive with this RT-qPCR assay were RNA sequenced at Medicinal Genomics with Illumina. Over 100M reads were sequenced from each sample. The viroid was detected in 8/8 samples at less than 1 read per million. Note the viroid has no PolyA tail so its capture efficiency in this process is likely very low but the sequence information confirms CAN1 and CAN2 have been sequenced confirmed with this assay.

DNA positive controls were serially diluted to construct a DNA based LOD. This was performed on Version 2 build of the MGC assay that has been optimized for Root extraction.

Hairpined CircRNA LODs are 2-3 CTs offset (later/less efficient) from their DNA LODs. Independent of this, the Version 2 assay is 10X more sensitive in Roots than our previous HpLVd assay. This is largely a result of the magnetic lysis step used to clean up dirty root samples.
Conclusions
Synthetically synthesized CircRNA can be injected into the stem of Cannabis plants at 723 Billion to 7.23 Trillion molecules and 10 days later demonstrate a linear to circular RNA difference in Cq scores with RT-qPCR and RNaseR treatment. Given these CT scores are lower (more RNA) than the original CT scores detected in the roots at 5dpi and the linear to circular RNA ratio is altered compared to the roots, this is highly suggestive of replication of the synthetic genome post infection. This partially supports Koch’s postulates as this synthetically synthesized and pure viroid is transmissible and replicative.
What has yet to be determined is if this transmissible viroid also confers disease in these cultivars as they have not matured to final flowering. The association with low yield has been made many times before but in studies that could not account for the vast array of other pathogens that may have co-associated with the transmission event. A very informative read on this topic recently published from Zamir Punja which demonstrates the pathogenicity of HpLVd appears to be cannabis cultivar dependent.
Final Sanger sequencing of the plant replicated viroid will be performed to verify the fidelity of the primers. Previous Sanger sequencing of viroids detected with these primers have provided 99% identical HpLVd sequences known as CAN1 and CAN2. These sequencing results are public on Viropedia.net.
The qPCR assay has been validated with Illumina RNA-Seq and can detect HpLVd viroids at less than 1 read per million of the mRNA in the sample (Billions of reads surveyed) or 5.5 copies per reaction.
The LODs for this assay has been calibrated on circular RNA templates and synthetic DNA templates. Since synthetic DNA templates are not hairpins and do not require a low temperature Reverse Transcriptase step, they provide an optimistic assessment of the true biological LOD. Synthetic DNA templates thus exaggerate your LOD 2-3CTs (10 fold), leading you to believe you have a much more sensitive assay than you really have once deployed on real circRNA templates.
For samples that are actively replicating and have more linear RNA than circRNA this distinction may be irrelevant as the linear RNA is not expected to be as difficult to denature and amplify as the circRNA. For latent samples where the RNA is mostly circular this difference may affect the assessment of the sensitivity of the test. As a result of the sequencing depth and synthetic RNA controls, we believe this is the highest sensitivity test on the market for HpLVd.
Understanding the viroids latency and replicative state may assist in breeding for cannabis plants more tolerant to HpLVd infection.
Some of the details on the RNaseR methods below-













Nothing will satisfy the "virus doesn't exist" mob. This even includes some who are very well known in exposing medical disinformation (generally on the side of truth). None of these aggressive loud-mouths have any knowledge of virology or molecular biology. Lack of knowledge is understandable and not in itself a "sin". Rudely and hatefully attacking those who do have the knowledge (earned through decades of producing original science data, analysis, and publication) shows severely flawed character. Those screaming the loudest, those hurling insults and worse, have no clue what is a virus, how viruses replicate, how PCR and sequencing function, etc. Nobody knows everything. I don't. When someone reveals information of which I am not knowledgeable, I take it in, evaluate it as best can, but never ruthlessly attack those who have extensive knowledge, making a fool of myself when I don't know what I am talking about.
I am not going to waste time and energy getting into a debate with the "no virus" mob. I have done this previously with very good intent, with the erroneous conviction that I could help them to understand the reality of viruses. This resulted in one of the mob leaders messaging me back, telling me to eff off. Another made false allegations on the internet and doxxed me. Then I put together a short explanation of only one of several lines of conclusive evidence for the proven existence and pathogenicity of viruses. Koch's Postulates (1884) have exceptions since scientific knowledge has grown so much during the past 140 years. I just checked and Wiki has a good summary of this. For any who may benefit I shall post one of several lines of evidence here. I do not wish to debate as it is akin to debating flat Earth, is frustrating, and pointless. No one has yet refuted this phylogenetic method.
UPDATED 11_15_23
“There is no virus” is a psyop, which seems intended to confuse, to “muddy the waters”, to divide, conquer, and discredit those who challenge the official narrative. The virus is real and infectious, and is not an influenza virus. Please note that the evidence I provide here does not indicate that a severe pandemic occurred. The evidence suggests it may not be more lethal than the flu. These issues are not the subject of this post.
PHYLOGENETIC TREES ARE PROOF POSITIVE OF THE EXISTENCE OF SARS-COV-2 VIRUS: The evidence presented here indicates only that SARS-CoV-2 does exist and my own personal experience is that the infection in some individuals is quite serious. It damn near killed me, did apparent permanent damage, severely sickened my daughter until I found her a doctor that prescribed ivermectin (she recovered in hours), severely sickened and damaged some friends. This is not “sniffles” as some have said. Easiest way to prove the virus—do an internet search for "phylogenetic tree of SARS-CoV-2". You will find that many independent research groups from many nations around the world have amplified, sequenced, and analyzed full viral nucleotide sequences of variants from tissues of patients and deceased, deposited these sequences in public data bases, and published analyses in peer reviewed journals. These are independent research groups using standard methodology and this cannot be faked (not on this global scale). I am a professional molecular phylogenecist and I have generated and published many such analyses over the years and taught this methodology to graduate students. For the most part, people who promote this "no virus" trick have never done this kind of work, which includes intense educational background, many years at the lab bench including PCR and nucleic acid sequencing, data analysis, and publication. Their misguided followers have never done this either. Those who say proof of virus is not a valid question have zero real world experience or comprehension in molecular phylogenetics, or any related science for that matter, and spread disinformation. Without comprehensive real-world, productive experience they cannot and will not knowledgeably address my point about proof of the virus using phylogenetic analyses. I will be glad to debate those with extensive expertise, but I cannot imagine that there is such a person, as anyone who fully understands what I am saying will agree with me. If you wish to challenge my comment, please begin with a valid refutation of my phylogenetic method of proof so as not to waste time and space advising me to read articles or visit a website, etc.
ISOLATION AND IDENTIFICATION OF SARS-COV-2 VIRUS: PCR primers isolate the viral genetic material. Transmission electron microscopy provides images. DNA sequencing identifies the isolated PCR product. Just as PCR and sequencing isolate and identify the viral genetic material, the same methodology is used in crime labs in every state, federal government, and around the world to isolate and identify genetic material from a contaminated crime scene to either exonerate or convict an accused suspect. In forensics, both direct nucleotide sequence and short tandem repeat (STR) analyses are used. STR analysis provides the number of tandem repeats at each locus, from which the nucleotide sequence is inferred. The validity of this methodology is established and accepted by the entire global scientific and legal system. If PCR and sequencing is reliable enough to isolate nucleic acid in order to identify a criminal perpetrator, the same method is also reliable enough to isolate and identify virus genetic material. If a critic does not accept this reality, then by default they do not accept forensic analysis. There are many convicts in prisons around the world that must be released if forensic methodology is flawed. If forensic methodology is not flawed, then virus genetic analysis must be accepted. It is the same methodology. One cannot have it both ways.