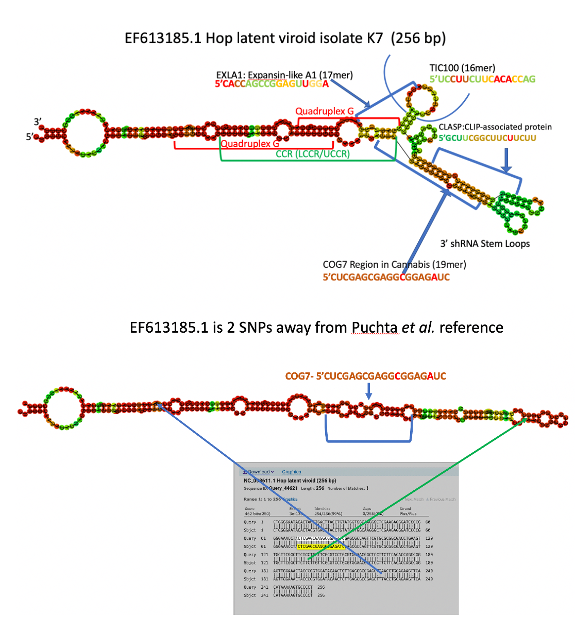
A viroid is wrecking the cannabis industry called Hop Latent Viroid. $4B in losses have been estimated. It’s mechanically transmitted and likely also transmitted in fungal mycelium, spores, maybe pollen and flies. It has reservoirs in other agricultural crops and it is a shapeshifter. The image above is what a Viroid cloud looks like. There are 162 genomes in NCBI for Hop Latent Viroid (HpLVd) and only 2 bases in this 256 nucleotide genome are invariant. Each non-colored base in the above picture is a SNP.
Why?
This 256 base viroid hijacks the plants cells replication machinery to make millions of copies of itself and it does so in the dirtiest possible manner. It hijacks RNA polymeraseII which is a DNA dependent RNA polymerase. This enzyme is used to making RNA from a DNA template (hence DNA dependent) and in the case of HpLVd, it is making RNA from an RNA template and making an error every ~500 bases during replication (estimated from PSTVd error rates).
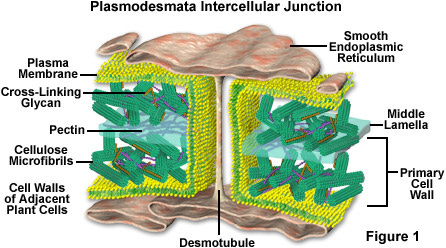
Viroids don’t code for any proteins. They just code for RNA that shuts down the expression of host genes with homologous sequence. In some cases these are Callose synthase genes which are responsible for regulating the porosity of cells and plasmodesmata. Once these are leaky the viroid can spread cell to cell.
Its’ error rate might be the reason why this viroid can jump from Hop to Cannabis and to Tomatoes. Its’ error rate is 10-100X higher than Coronaviruses as CVs code for their own RNA dependent RNA polymerase and in the case of SARs-CoV-2 they also have an ExoN gene that establishes proof-reading. This higher fidelity transcription is required for 29.9Kb RNA viruses. Small viroid genome sizes are likely a result of their high error rate.
This also means viroid clouds are more diverse than CV clouds and it is unlikely that a plant is ever infected with a single viroid genotype. More next gen sequencing is required to truly sort this out but this appears to be the case in other Pospiviroidae like PSTVd.
How does this thing do this? To start, it is a circular but mostly complementary genome (hairpin) and this circular genome enables some interesting amplification tricks we discuss more in our preprint. The latitude like lines on the graph are stretched hydrogen bonds and represent complementary bases.
The plants host RNAses (DICER) dice this genome up into 21-24bp dsRNA pieces. These pieces are loaded into RISC which targets host genes with similar sequences and digests them. The HpLVd RNA structures contain a two overlapping quadruplex Gs which appear highly conserved. These structural motifs are believed to create catalytic RNA or Ribozymes that target its own genome for digestion into 256 base pieces in hopes that the plants endogenous ligases will glue them back into circles and repeat the process. The replication process was already discussed on this thread.
With all of this diversity, the secondary structures in the RNA are also varied. Just 2 SNPs and the viroids structure changes dramatically.
There is a deeper dive on this topic on our recent preprint. You can explore this diversity on this recently built interactive Viropedia.net.
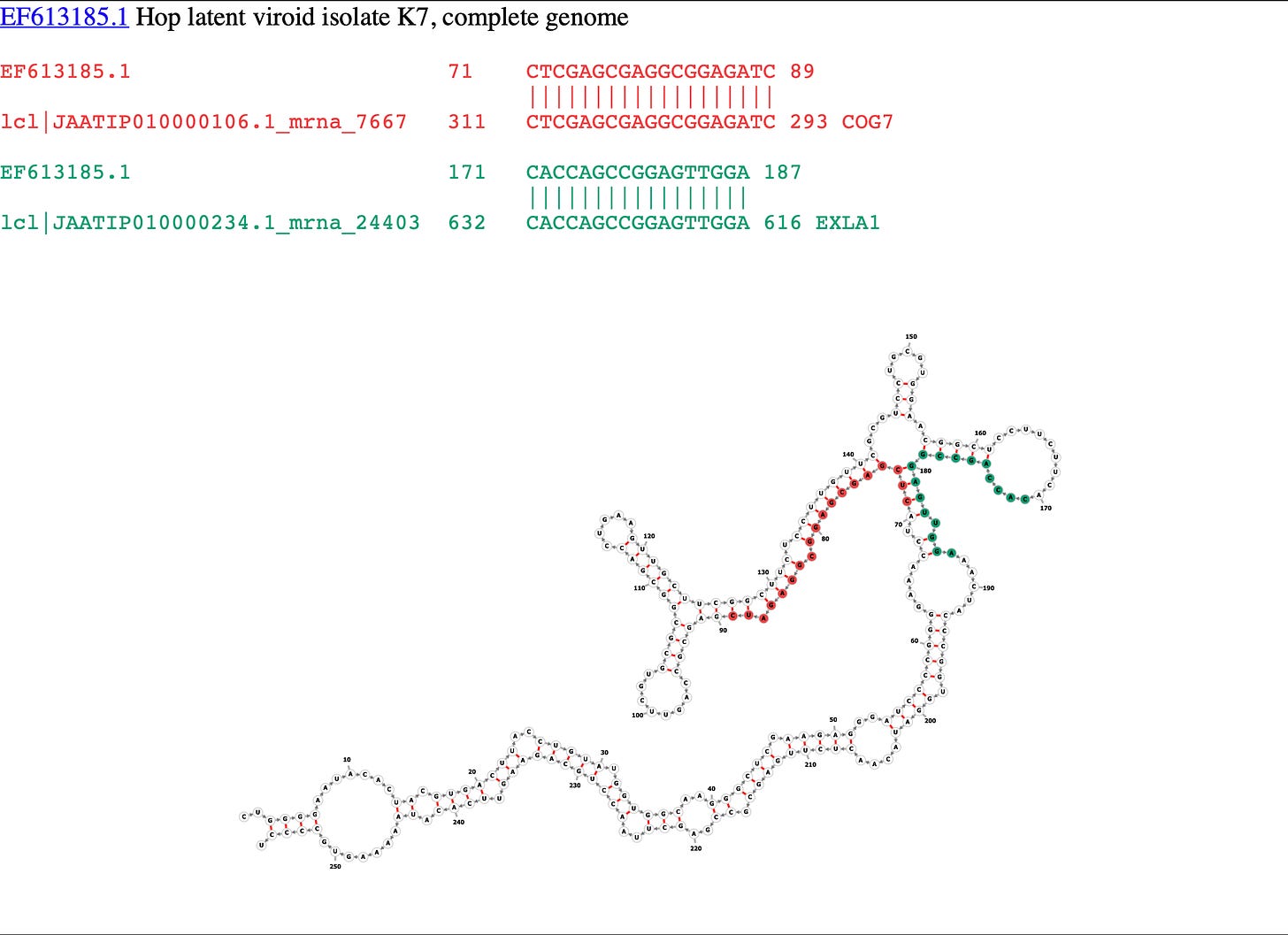
It won’t load on a phone yet so you’ll need a laptop to get a handle on this. This page decorates all of the HpLVd genomes known to date with their sequence homology to certain Cannabis mRNAs. There are 25 to page through. The website is currently only decorating the 3 more interesting targets (COG7, EXLA1 and CLASP). However, the more we dig on this, the Callose synthase (CALS8) gene and a few others will need to be painted on in time.
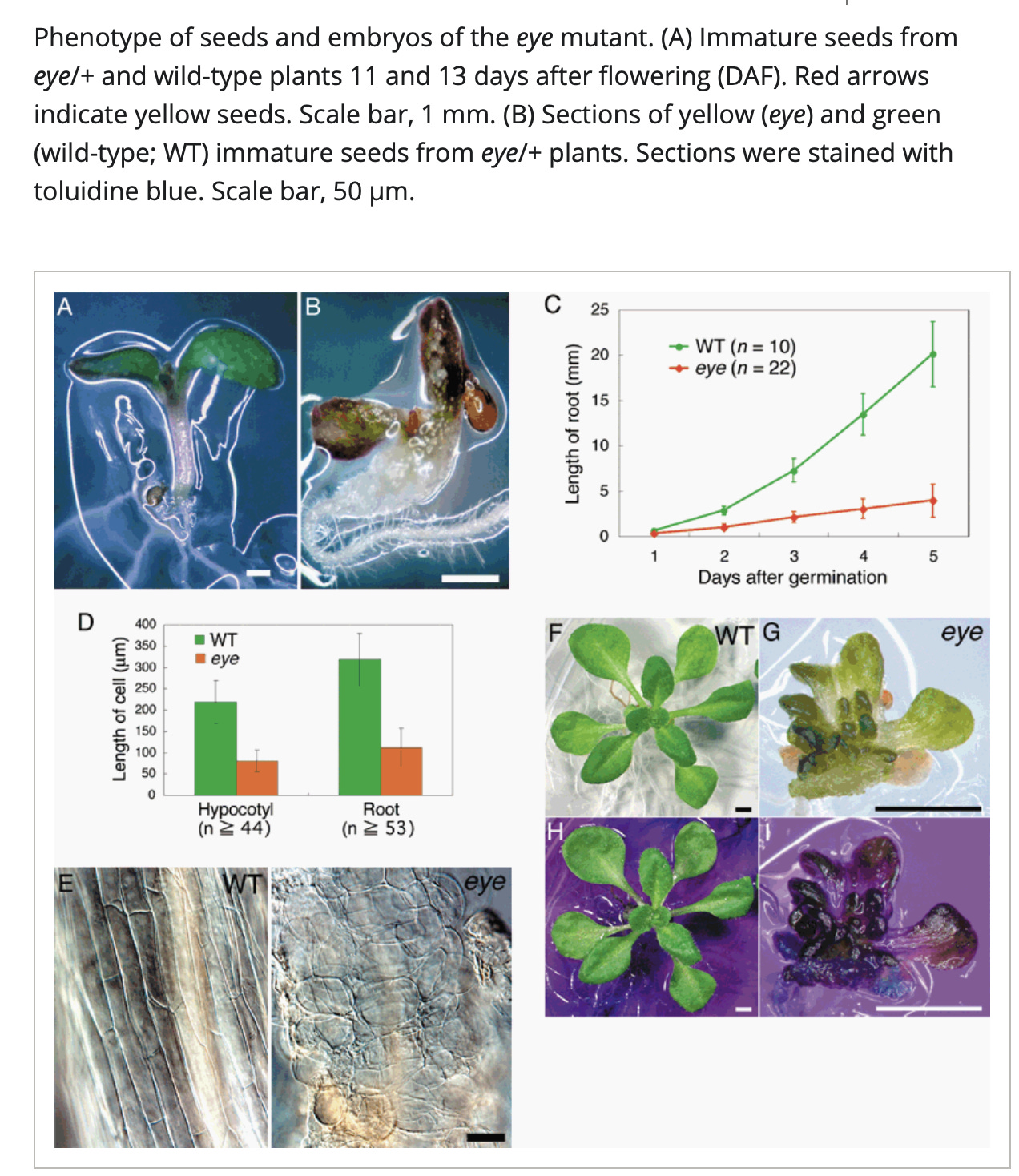
So what is COG7 and why do we care if that gene is down-regulated? COG7 Knock outs in Arabidopsis can provide some insights.
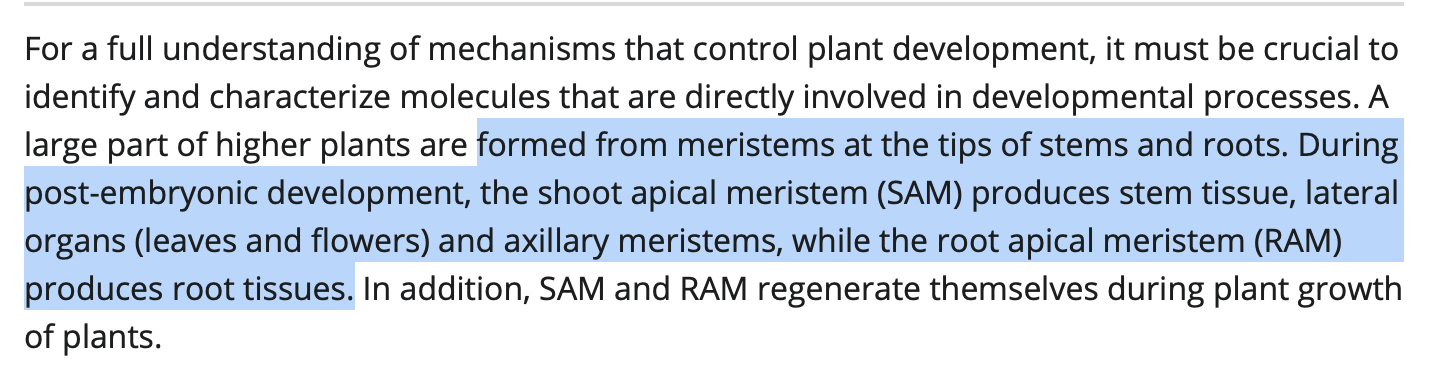
What does this look like in developing plants?
Leaf morphology goes haywire once the viroid comes out of latency and manifests disease in cannabis flowering. Meristem growth may also be impacted in cannabis infection.
A good review on a more studied viroid in the same Pospiviroidae family is focused on PSTVd (Potato Spindle Tuber viroid).
There are some complicated steps in this pathway. First the viroid genome is replicated (with errors) with endogenous plant DNA dependent RNA polymerases. These copies get diced up by an RNAse known as DICER. These RNAses make 21-24bp dsRNAs that are then loaded by RISC or the RNA induced silencing complex.
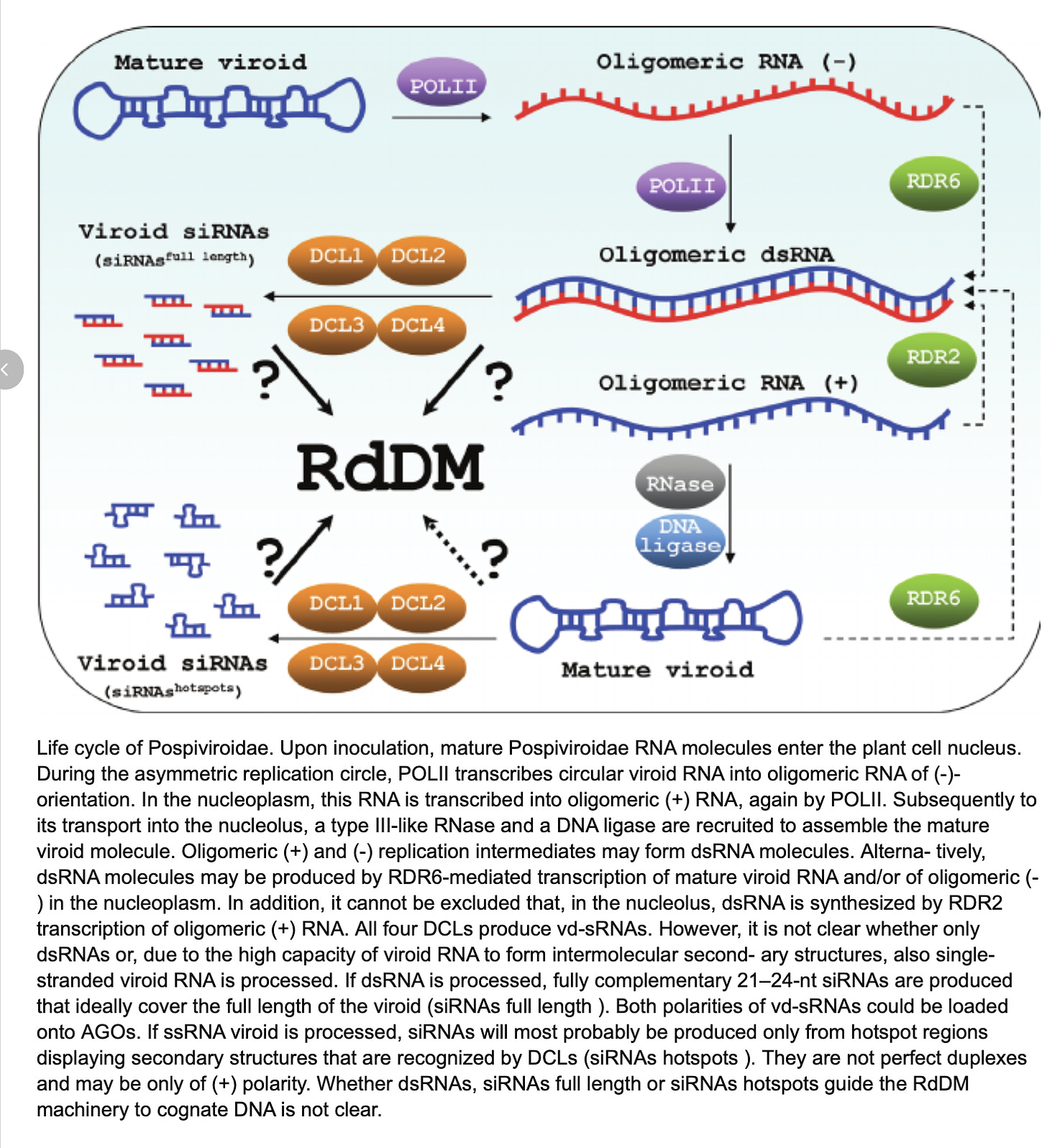
These RISC enzyme complexes (Argonaut) rely on a core of 8-9 nucleotides to have exact matches to their target genomes and are more tolerant of 21-24mers that only have 19 bases of homology to Cannabis. Adkar-Purushothama et al are some of the pioneers of this space known as vd-sRNAs. These are viroid derived small RNAs. Viroids don’t infect mammals but there is evidence of them infecting plants and fungi.
Table 4. above shows the error tolerance in the dsRNA when hybridizing to the hosts mRNA. So while the cannabis-like sequences found in HpLVd are short, DICER will still chop these up into 21-24mers of which 19-24 of the bases in HpLVd will be near exact matches and likely enable RISC to down regulate about 25 different genes. This of course, needs to be verified with some RNA-Seq experiments underway.
The PrePrint has this gem of a table that spells out all of these genes and lists a few papers that support various hits.
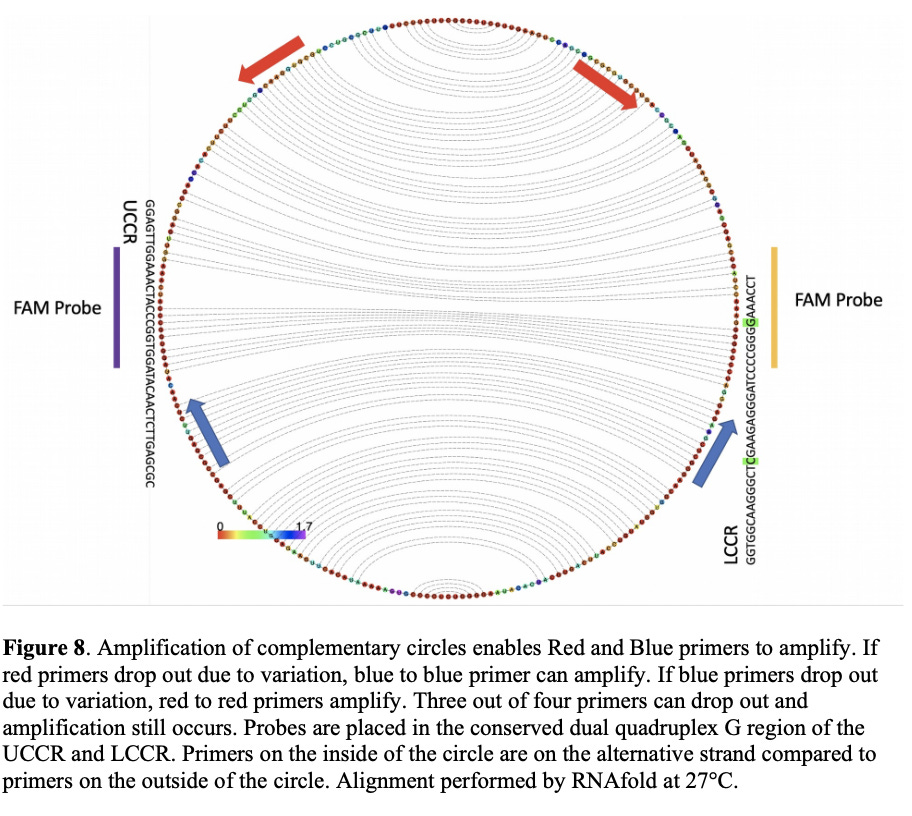
Due to the nature of the sequencing done to date, about 40 bases of the 256bp genome is under sampled. This is due to most methods using PCR to amplify these genomes and sequence them. PCR needs 2 primers (20bp each ) to amplify these circles and the sequence derived from primer sites will always match what the oligo synthesis shop synthesized for you as the polymerase builds templates off of this synthetic piece of DNA. Therefore SNPs cant be ascertained in primer sites.
Newer methods are under development that will amplify this genome as 2 circles and resolve these under studied regions.
So what do we do with this information? Do we fight it harder with a Zero COVID like approach knowing the viroid is more mutagenic, smaller, has no protein coat, can travel through fungal networks and spores and infects many different kinds of plants?
I thought Zero COVID was an irrational. An intentional dismissal of any type of cost benefit analysis that all businesses need to constantly be evaluating. I think its even more unrealistic to think the cannabis industry will suddenly eliminate HpLVd when it was discovered in 1988 and its still here.
While the PCR testing treadmill is great for PCR testing companies like ours, we feel obligated to chart our own obsolescence. This has been our experience in genomics. If you are not actively looking for better ways your customers could be spending their money, someone else will be and its better to be embracing this charge with your customers than chasing it.
Identification of the potential HpLVd targets in Cannabis leads us on a hunt for cultivars that have SNPs in these regions. Kannapedia.net in fact has 5. Breeding with these may produce plants that are less symptomatic with HpLVd.
Lessons from COVID PCR: CT scores matter and PCR positive asymptomatic plants are valuable to study. This is the conundrum with HpLVd PCR. Cannabis mutations that halt the RNAi, wont stop the replication of the viroid and aggressive programs that cull every positive plant without CT nuance or COG7 tracking, may in fact cull the very plants more likely to breed us out of this mess.
We need to be honest about the likelihood of an imminent cure for this. Many published approaches for viroid eradication have been proposed. Some are getting into the complicated space of Plant Growth Regulator regulations and no one wants absorb transmission risks trying to sort our cures. There is too much at stake to be fiddling with this in production grows. Isolated breeding efforts or tissue culture labs are needed to breed the next line of cannabis strains that don’t suffer yield losses upon infection.
It is a longer term play but what one learns on the journey will be additive to a grows over intelligence and growth of their operations.







1) Thank you, but this post is too difficult for amateurs to read, it makes our brains hurt. This should be published in an obscure journal where only a thousand of your peers will have to see it!
2) You note that viroids don't infect mammals, so I suspect your cat is safe. If you could explore the mammalian mechanism of resisting viroids you might find a useful pathway (other than Burbank methods) to protect your grow-ops.
3) Would your lab potentially be able to take samples from 10 vaxxed people and ten unvaxxed and determine the existence of spike transfection into DNA in (human) vivo? The vaxxed should all have Wuhan spike incorporated (somewhere), while the naturally infected (at least 90% of us) may have Wuhan spike, any Greek letter spike, or preferably no incorporation. Lymphocytes from blood donors might be an accessible source, and you could also have your grad student or fellow look at the T-cells to confirm immune status.
Crazy mutations in the photos. Do the mutations appear similar in other species, say tomatoes?