There is a viroid that is a Billion dollar wrecking ball to the Beer and Cannabis industry known as Hop Latent Viroid (HpLVd). It is spreading through the US, Canada and Israel and results in stunted growth and low yields. The industry is reflexing to ever more sensitive qPCR tests and culling everything that has a shred of qPCR signal. Very little sequence validation exists for these late CT ‘Positives’. Cyclers are cranked out to 50 cycles, multiple amplicons are being pooled to target a 256bp viroid, and in many cases 50% of all plants under lights are getting culled.
I understand the reflexive response when so much is at stake but it is very likely going to result in culling the very genetics that may get us out of this mess. This happened in the C19 pandemic. Panic set in and testing companies spread fear while bragging about their single molecule sensitive tests.
Boasting about a sub 5 copy sensitive qPCR test often just highlights ones lack of understanding of the principle of digital PCR and what stochastic sand such claims rest on. At best these tests will be fractionally positive due to poisson sampling of such low amounts of DNA/RNA. For those not familiar with digital PCR, its the process that most Next Gen sequencers rest on. When you dilute your DNA down to such low dilutions such as a single copy/ul, when you pipette a ul, you dont always get 1 copy. Sometimes you get zero and sometimes you 2 copies. Poisson sampling runs the show at these dilutions and results go digital.
So pushing your tests into this territory, while impressive, is often a misguided brag as you cant rely on the test to always be positive at such dilutions as Poisson will not allow for it.
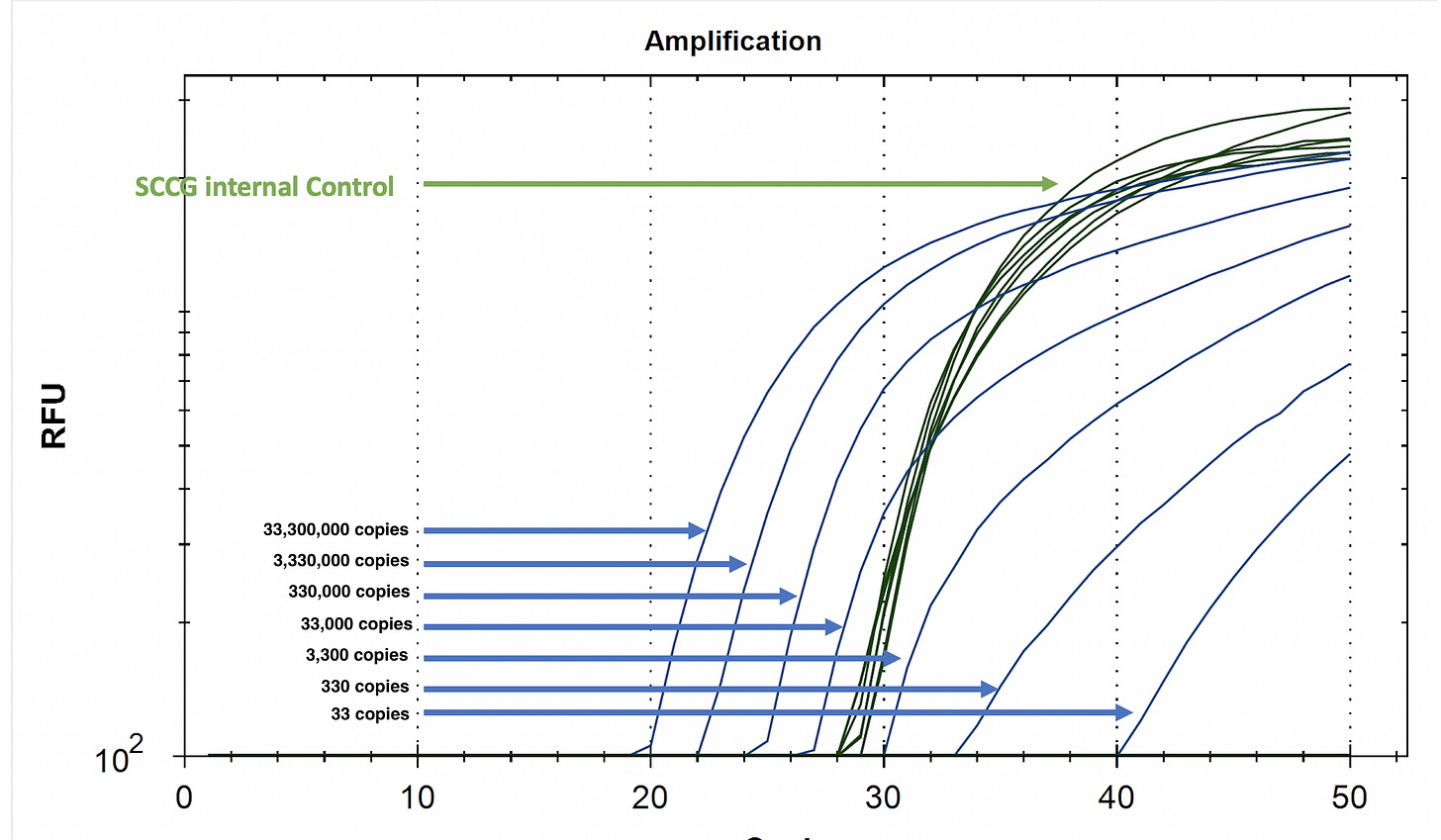
Included below is a Limit of Detection (LOD) of a Multiplex HpLVd assay we have on the market. It has two amplicons targeting the 256bp viroid both labelled in FAM. The two amplicons are needed to dance around SNPs in this quasi-species. It has a 3rd assay targeting an expressed cannabis gene as an internal control (Green in HEX) to ensure the leaf lysis performs. This is critical with fast boil preps where no DNA/RNA purification is being performed as fast boil preps are notorious for qPCR inhibition and you need the internal control to signal when a sample is truly negative or just inhibited from a foliar spray that may be choking the PCR reaction.
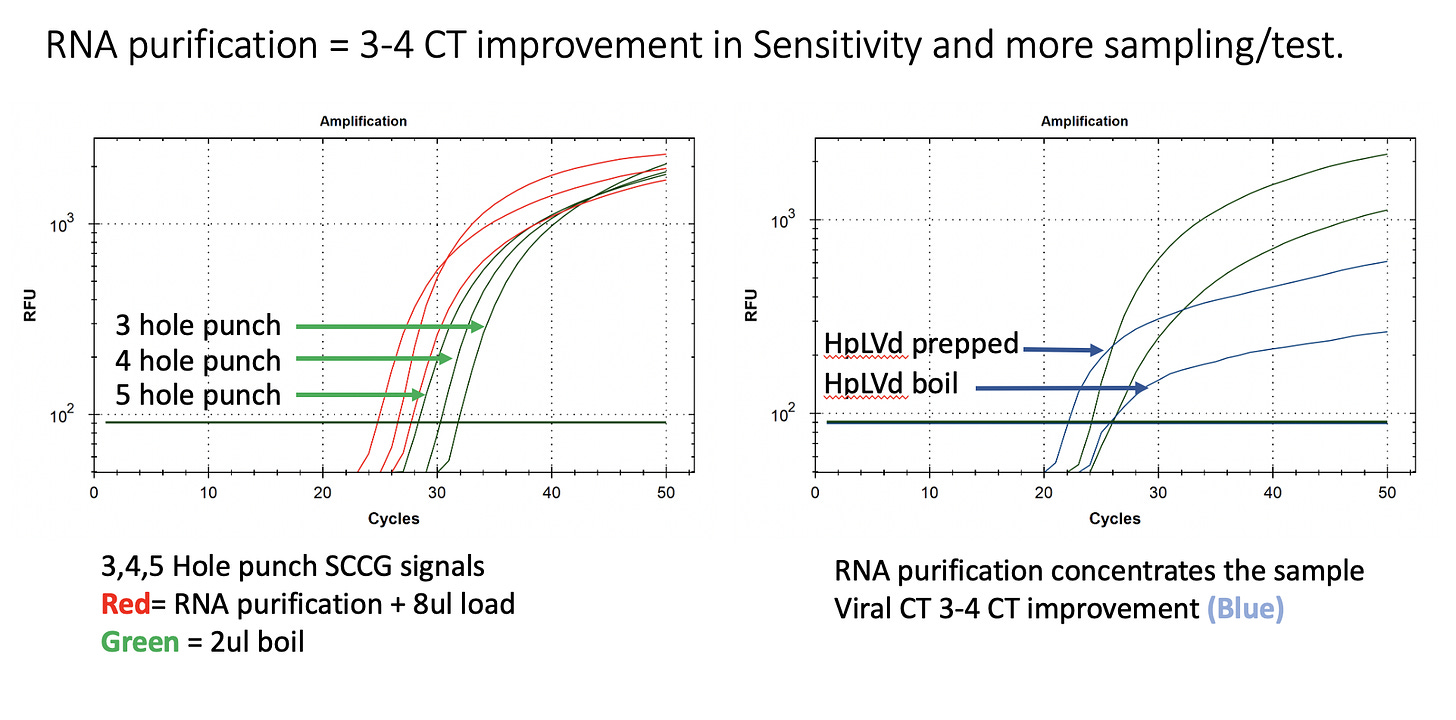
This test uses 2ul/50ul total lysate and the PCR reaction can actually fit 8ul/50ul of lysate improving the LOD to below 9 copies. Concentrating the RNA with a purification step brings this assay into single copy territory but buyer be warned!
One could certainly optimize this even lower with a RNA purification that concentrates the RNA of all 50ul lysate into 8ul but at some point you need to back up and consider the corner you are painting yourself into. qPCR below 5 copies ventures into territory that is extremely expensive to sequence verify. You are operating in the technologies least credible performance window where digital PCR artifacts plague the sampling and False positives/False negative escalate exponentially.
It is better to operate qPCR in a range where these technical artifacts are not at play and sample more tissue to compensate. Surveying more of the plant is going to provide better assurances than hyper sensitive tests on a single punch.
Most other single CFU assays (Aspergillus, E.coli, Salmonella) in the cannabis space rely on enrichments to compensate for various subsampling artifacts and help to move the qPCR assay into a window where were are measuring 100s of molecules to make a single CFU call. Viroids don’t culture or enrich leaving us to measure them directly.
What we learned from C19, is that viruses have certain lifecycles where they are making replication competent genomes and where they are being defeated by the host and the presence of their RNA fragments still persist but are not infectious. Many people can be PCR positive 90 days after they clear the virus and people who are immune to the virus, still become PCR positive but present less symptoms. Presence of RNA is not a sign of infectiousness and if immune plants can still have the presence of this RNA. We need to be aware of any culling procedures that might cull the resistant cultivars.
Keep in mind this is a viroid that significantly impacts flowering (and cannabis reproduction) and thus is swimming upstream in an evolutionary gradient.
It is tempting to not split these CT hairs in cannabis and just cull everything that has any signs of HpLVd RNA even if testing companies offer no evidence of sequence validation from their late CT tests. This is an understandable approach with disposable clones but may be catastrophic if applied to valuable mother plants.
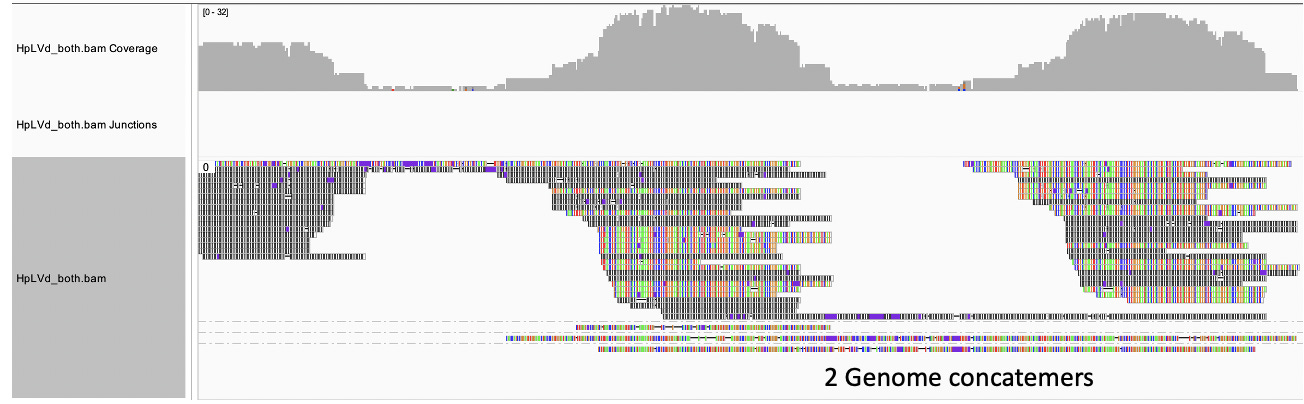
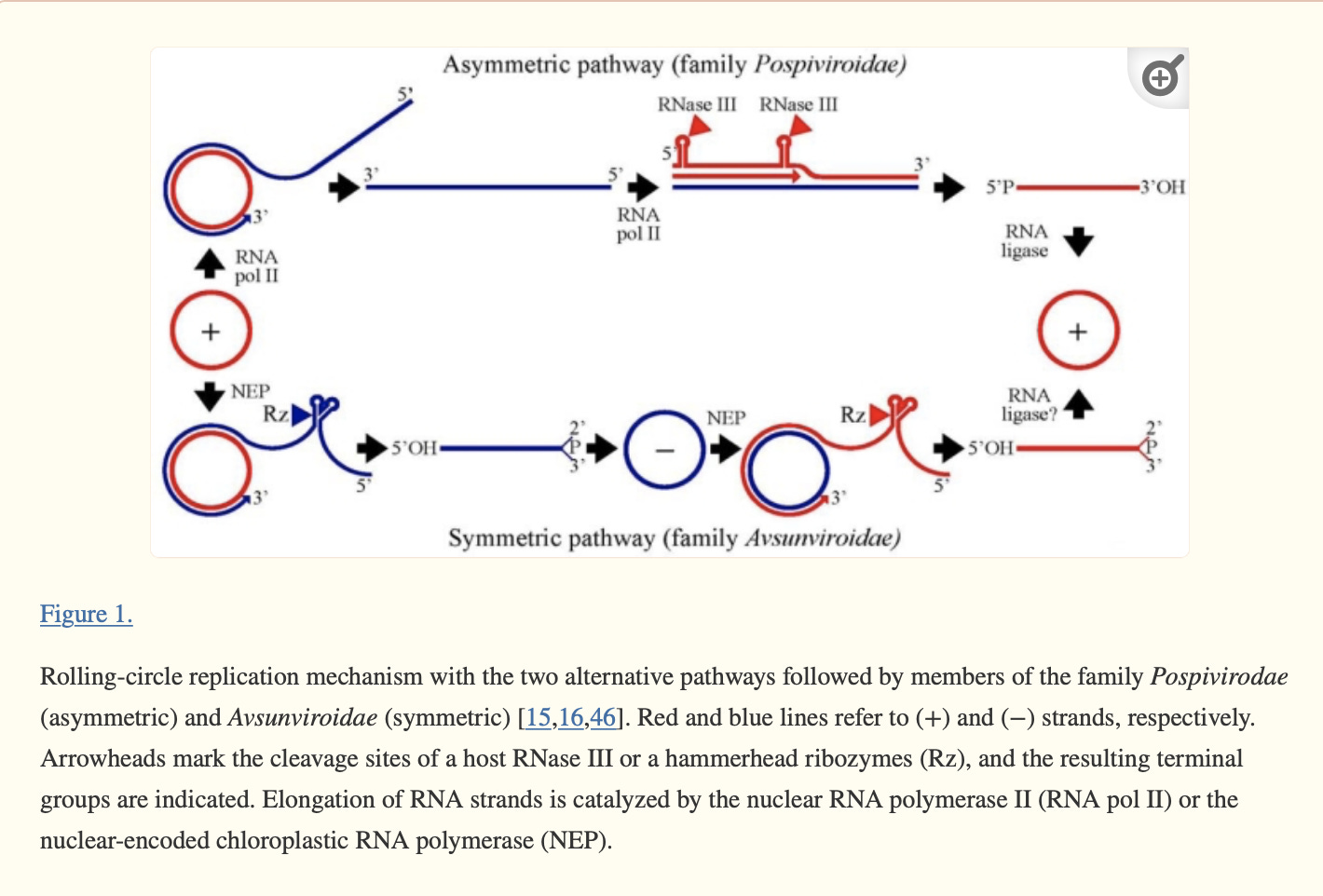
Let’s review the life cycle of Viroids and see if there are parallels to C19. Much of the literature is focused on viroids in the same family as HpLVd but doesn’t explicitly cover HpLVd so there is some extrapolation going on. The collection of evidence points to HpLVd being circular (more below on this) and relying on Rolling Circle amplification to make copies of its genome. The circle is Replication competent. The linear copies of its genome ARE NOT replication competent but will provide qPCR signal. The circular RNA is also more stable than the linear RNA. These linear copies require further processing to become circles again and various activities of the plant may inhibit these steps. For instance, if RNase III is suppressed, the linear genomic concatamers cannot be processed into replication competent circles. If the Ribozyme or RNA ligase is suppressed (they might be in veg stage), you will have circles that make linear concatemers but fail to make more circles.
RNase III inhibitors have since become an intense area of focus for addressing plant related virus problems. Suppression of RNase III is believed to leave the viral (not viroid) genomes more intact and thus targets for the natural viral defense mechanisms. You can think of RNase III as a tool that chops the viroids up into pieces too small for the immune system to see but still large enough (21bp) to play a roll in RNA silencing via siRNAs. RNA interferance is the predominant theory as to the pathology of HpLVd. RNase III or the HpLVd’s own Ribozyme activity digests its RNA into short interfering RNAs that create havoc in RNA regulation.
Many plants express RNase III as defense against viruses. This is great place to look for variants that might confer resistance to HpLVd.
Is it a circle?
But before we jump to Cannabis RNaseIII genes….
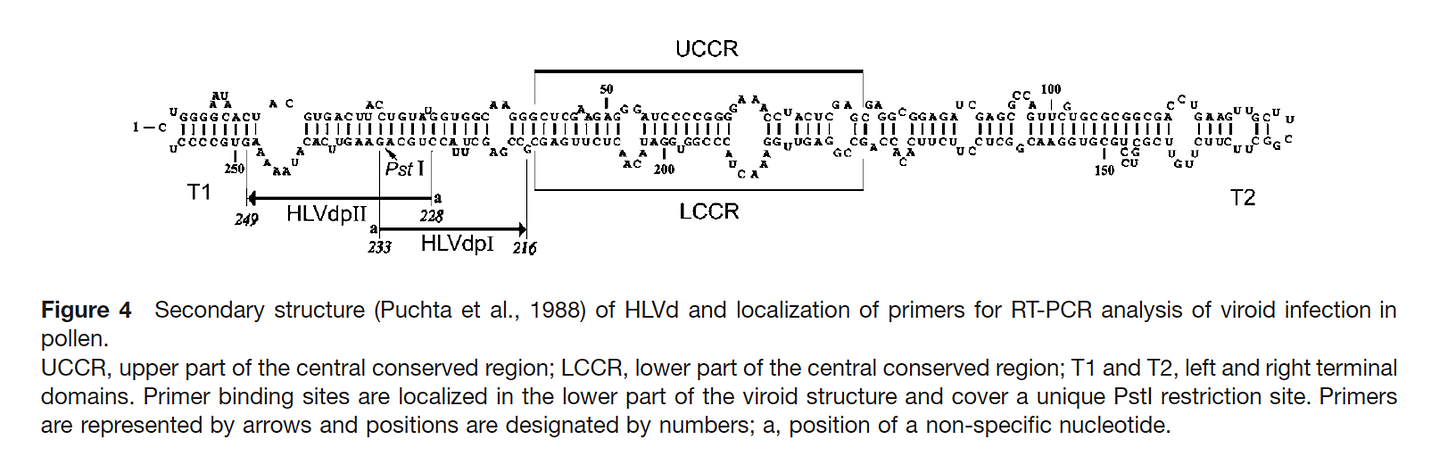
We should confirm a few assumptions. A few people in the field were questioning the circularity of the HpLVd genome and since the papers describing this were published in 1988 (Puchta et al) , surely there must be better tools to measure this today.
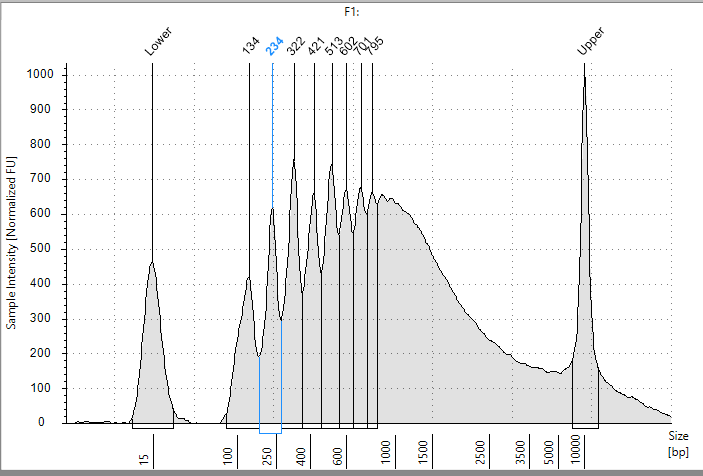
So we did a simple experiment. Bst Polymerase is a strand displacing polymerase with RT activity. 8hr polymerization with Bst Polymerase primed from a single primer (65C) from our PCR assay should deliver us a single 256 base pair band if the viroid is linear, or a concatemer if its circular.
This above Agilent trace certainly shows signs of concatemers and circularity but we were not expecting polymerase stall points at every half turn of the viroid (shown by the peaks 134, 234, 322, 421 etc..).
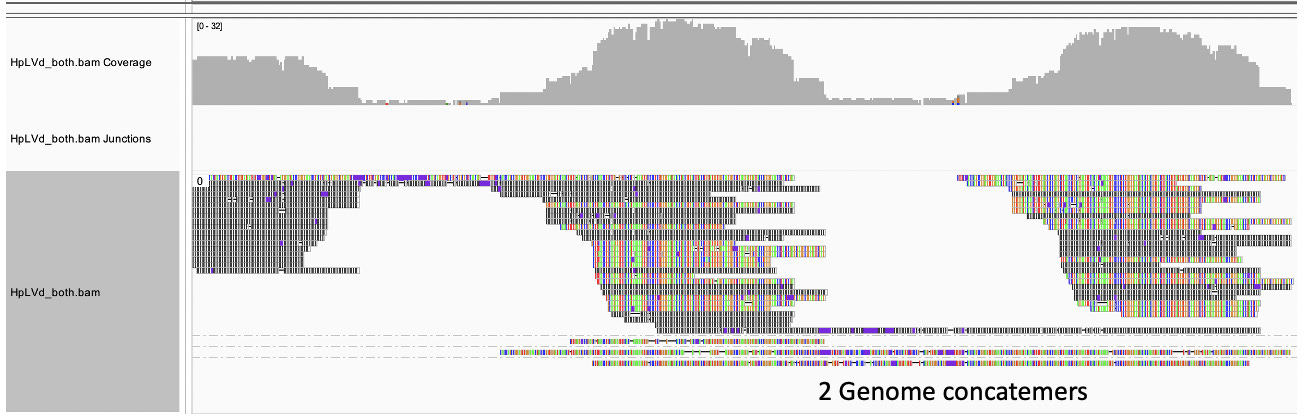
To further understand this we sequenced these Bst polymerase concatemers with Oxford Nanopore Flongles. This was a bit tricky as ONT doesn’t like long hairpins. These seem to clog the pores and provide lower quality sequence.
Nevertheless, we can see several reads when mapped to a concatenated reference, span multiple HpLVd genomes but there is a reduction of sequence coverage in certain hair-pined regions of the genome. This supports the notion of the genome being circular.
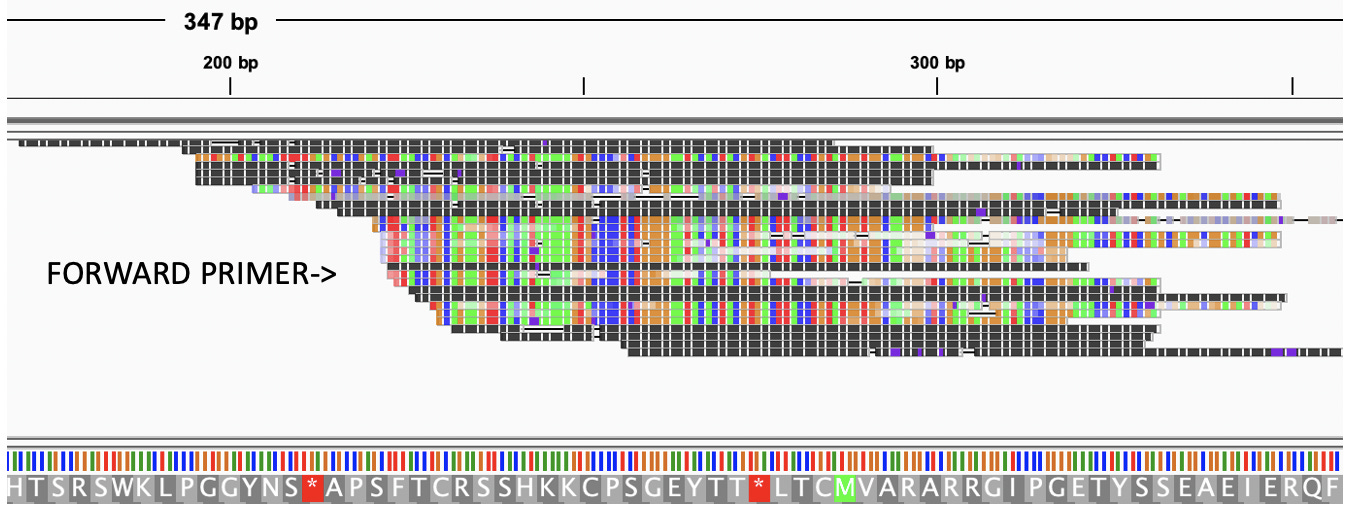
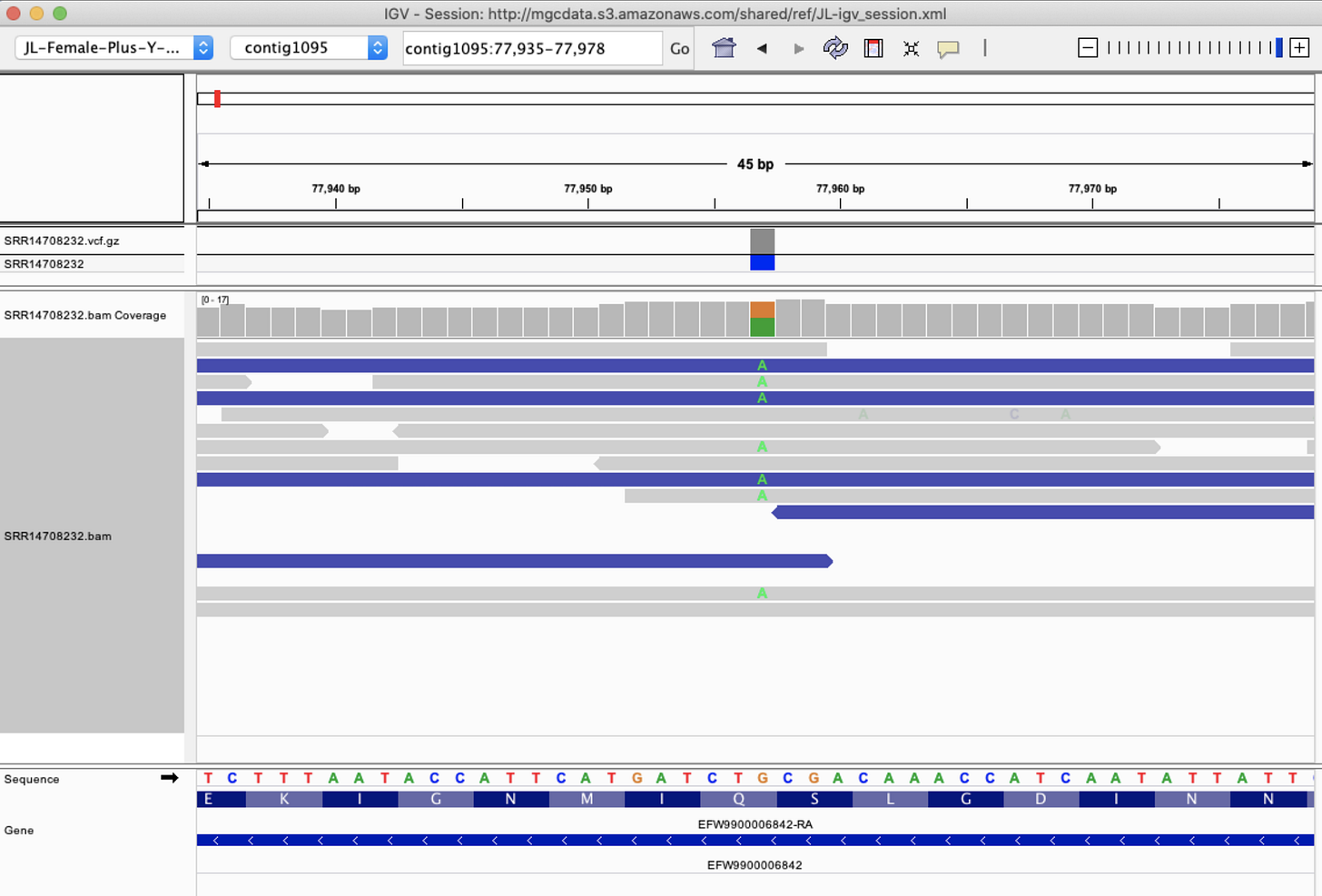
So does Cannabis have an RNase III gene?
Jamaican Lion does-
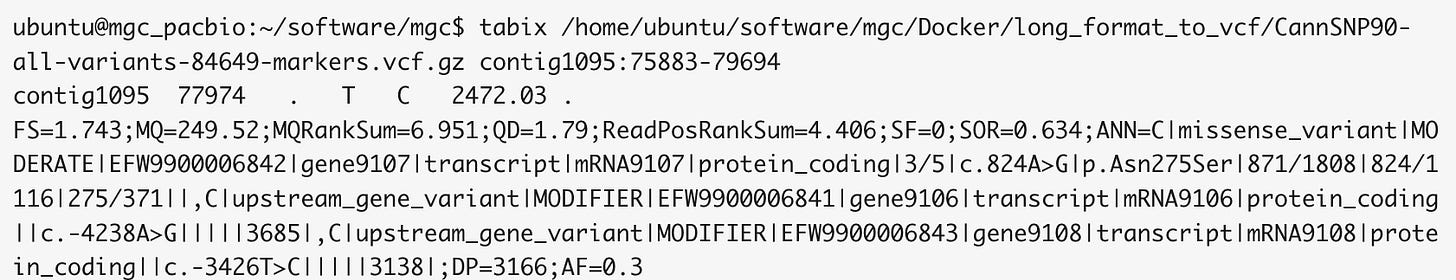
And according to our CannSNP90 data, there are variants in this gene that might make sense to track in your plants.
If you look through all of the data in Kannapedia.net which includes whole genome shotgun data, you find even more.
Some cannabis genes in the neighbor to RTL2 in the cannabis genome are Auxin-responsive proteins.
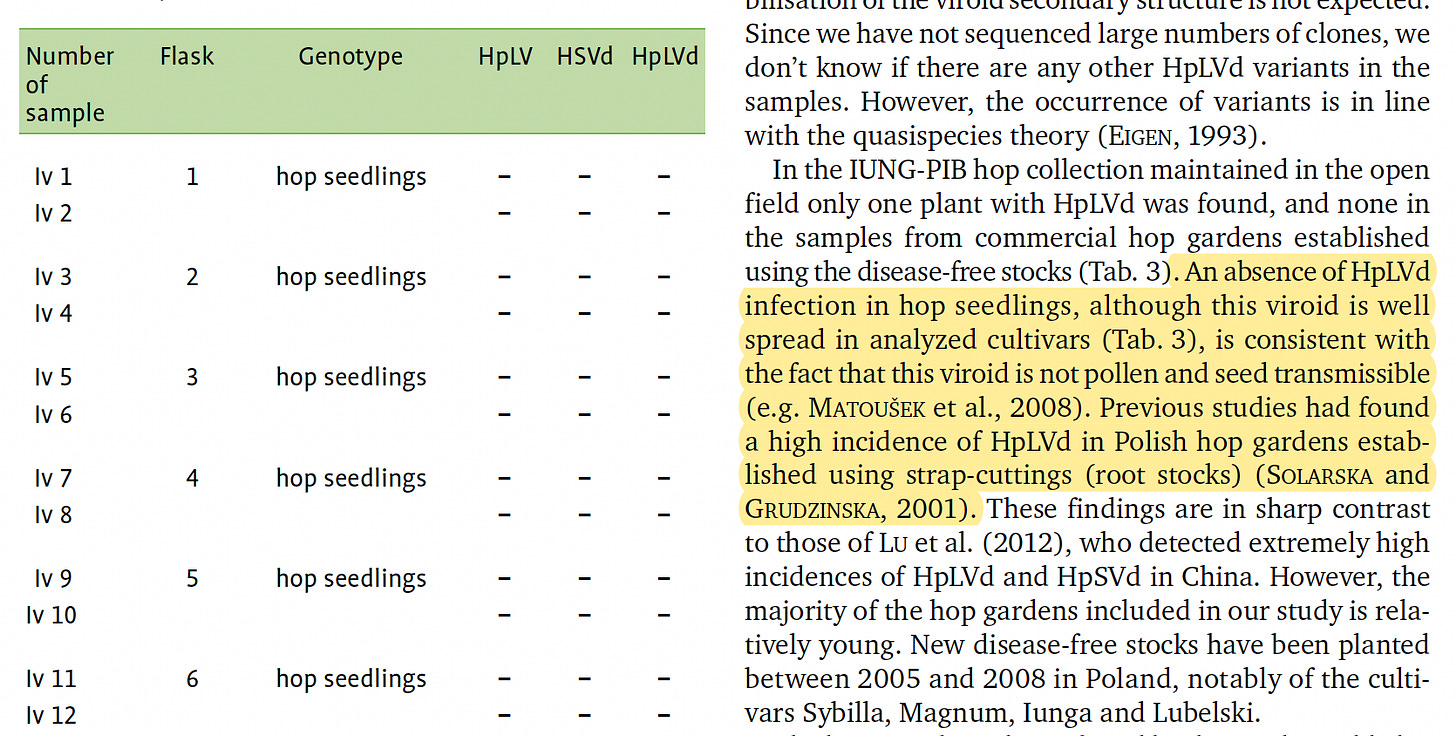
These nucleases are one of the reasons the literature suggests HpLVd doesn’t transmit well in Hop seeds. Pollen expresses nucleases to ensure a clean generation.
This work was performed in Hop and may present different results in Cannabis (Ziegler et al). When assessing seeds with qPCR for HpLVd, it will be important to first bleach the outside of the seed before grinding them up for testing. The seed coat is maternal and may contain remnants of the viroid that is unlikely to survive the germination phase.
Why am I bringing all of this up?
Plants that fight off this viroid are likely to be qPCR positive but later CT. It would make sense to sequence any plant that is positive but doesn’t show signs of symptoms upon light cycle changes. Breeding with those is your best shot at getting around this longer term, otherwise your qPCR viroid whack-a-mole will never end.
Also keep note of the CT scores. The plants with replication competent circular viroids, tend to have CT scores in the 15-25 range. The CTs in the 35+ range are very likely non infectious RNA fragments, false positives or contamination.
It is better to survey more tissue than to chase single copy sensitive tests. We are exploring pooling many hole punches into a single boil. This will likely require a RNA purification step to manage this much tissue but it will also concentrate the sample improving your sensitivity in a more robust and reliable manner than squinting at CTs past cycle 40.
These are 150ul boils. After which 100ul can be collected and RNA prepped in a deep well plate. Eluted in 10ul and you have 10X the concentration surveyed in 3-5X more tissue.
Some people take approaches this like and convert them into a pooling strategy where they pool the hole punches from 5 different plants in a fertigation row and if any of the rows are hot, they then retest at an individual level to find the hot plant. Pooling strategies make sense when the population frequency of the disease is below your pooling depth, such that most pools are negative and you are only retesting an occasional row. Once the disease prevalence is higher than your pool depth (1/5 in this case) then every pool is positive and you have to test every plant anyway.
Using this multiple hole punch/test approach for surveys, increases sensitivity without pushing the qPCR test into its zone of unreliability. While the purification is an additional 30 minutes of work, it is magnetic bead based and is easily automated on a variety of liquid handling instruments we support and help to program.
In summary, there are many reasons to adopt genomics to eliminate HpLVd. Proper use of qPCR can avoid chasing RNA ghosts. Scrupulous use of CT scores is required to eliminate infected clones and perhaps hunt for asymptomatic carrier plants. Sequencing such plants might reveal important markers for HpLVd resistance. Sequencing the HpLVd genome in your facility may aid in tracking the epidemiology of the disease.
Contact us at Medicinal Genomics if you are interested in sequencing HpLVd genomes or cannabis genomes. We can also help craft strategies that help to test for this viroid and eliminate it from your grow with minimal FPs and destruction to your crop.











Kevin- you wrote an article showing how PCR tests are used correctly in your research and why the PCR test did NOT work to detect CVD reliably, but I cannot find it. Can you send it to me? Khenson@sedgpc.net. Thanks!