Sequencing of RNase A treated Pfizer bivalent vaccines reveals paired-end sequencing evidence of circular plasmids and an inter-vial 72bp variation in the SV40 promoter.
Introduction
Previous work sequencing the Pfizer bivalent vaccines utilized a method that sequenced both the RNA and DNA in the sample. The RNA-Seq kit employed favored sequencing RNA and under estimated the DNA contamination levels in the vaccine. This made it difficult to assess the % of the plasmid DNA that was linear versus circular DNA. To address this problem, we RNase A treated the sample and sequenced the remaining DNA. RNase A digests RNA into small pieces that can be eliminated with purification. This enabled a deeper sampling of the DNA based molecules without the mRNA sequence obscuring and suppressing the depth of coverage for the plasmid vector.
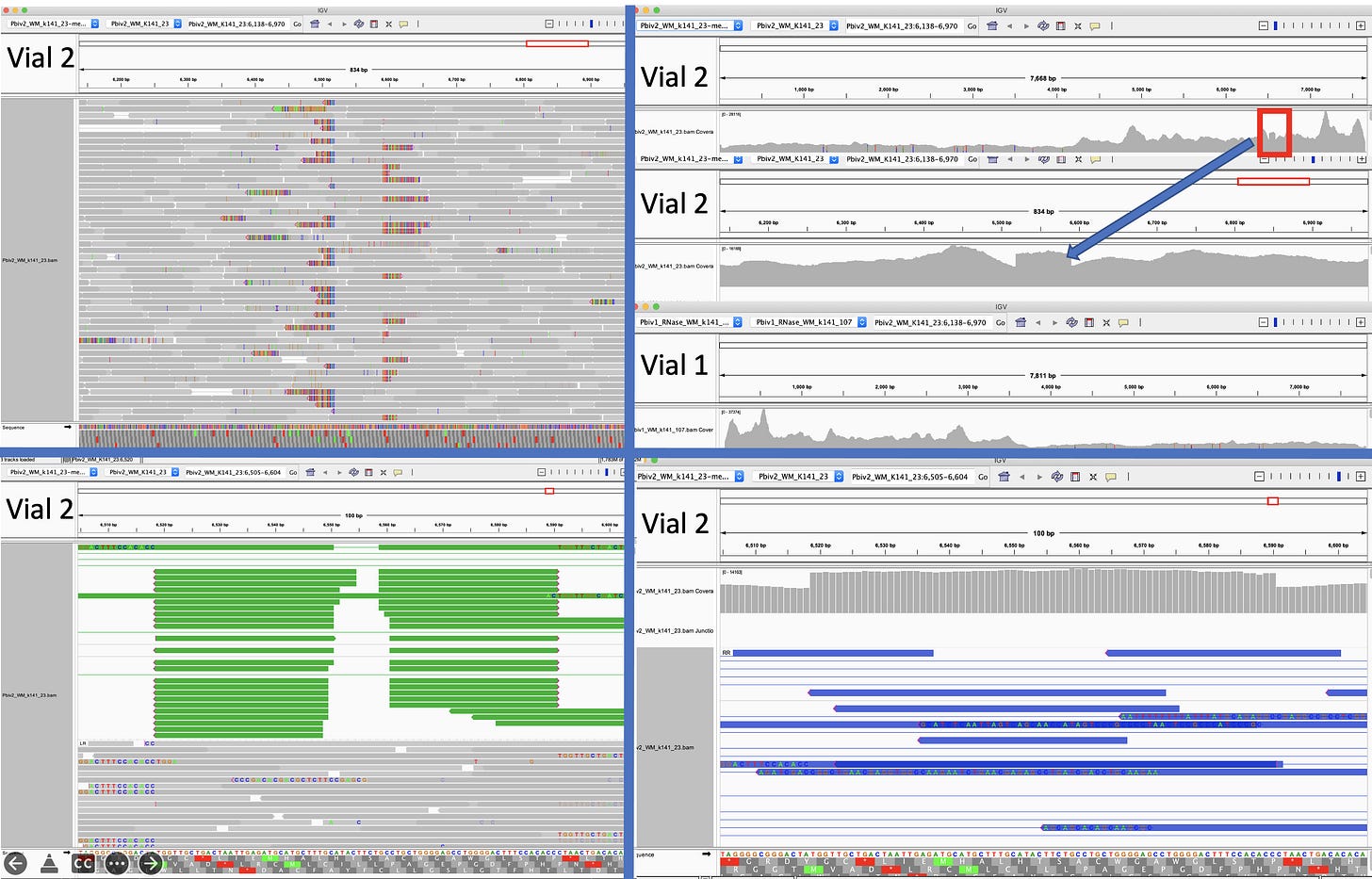
After assembling and mapping these paired reads back to the reference genome we can look for reads that point across the part of the reference that should be circular. Since these sequencing libraries were known to be 250 base pairs in size, we can look for reads that map ~7,700 bases apart and point across the circularization point.
Let us expand on this point as it a very important aspect of next generation sequencing. Figure 2 is the DNA length of the RNase A treated libraries. We were very careful to make these very tight insert size libraries so we have an expected distance between the forward and reverse sequencing reads. The tighter your size distribution of molecules, the easier it is to detect deviations from that distribution.
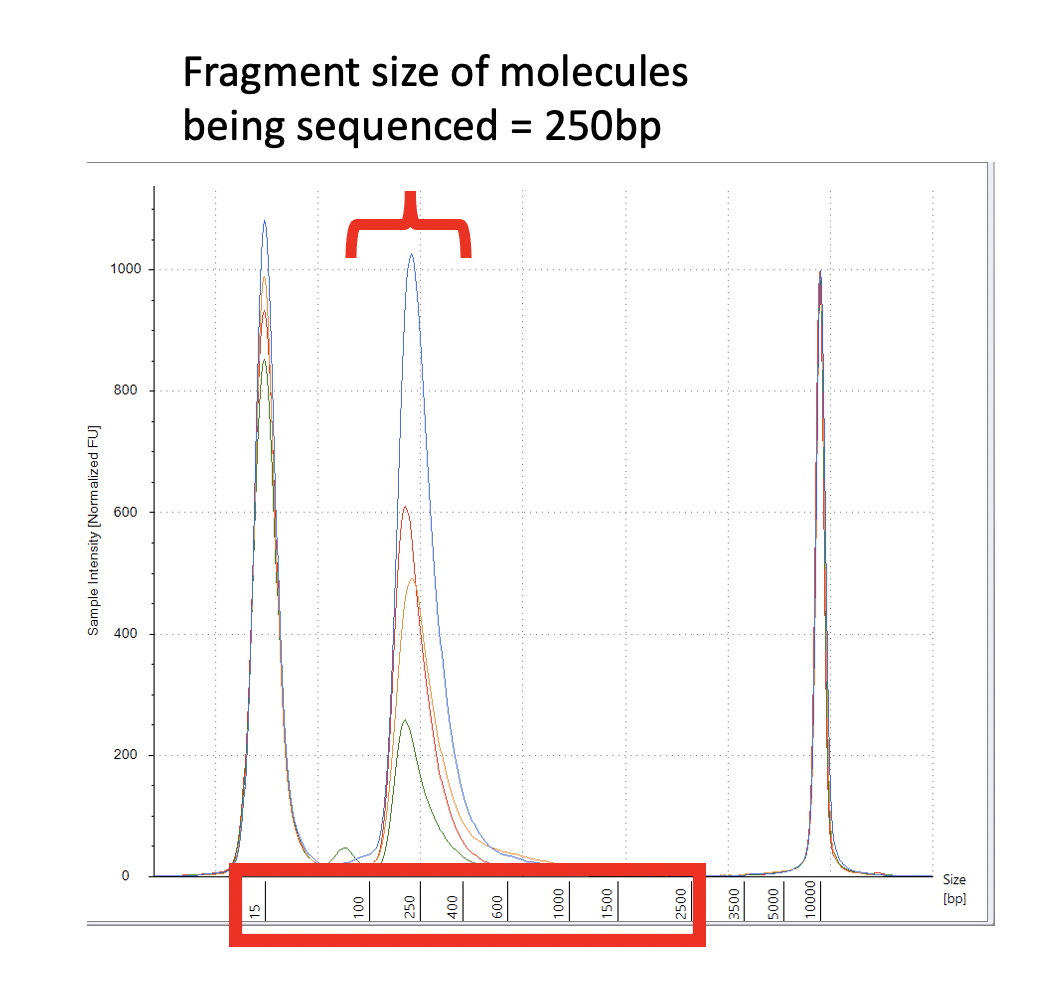
When the reads map to the reference and significantly deviate from this known DNA length, you know you have structural variations in your molecules compared to the reference genome you are mapping them too. NCBI has a tutorial on how to detect structural variation with short reads (Figure 3).
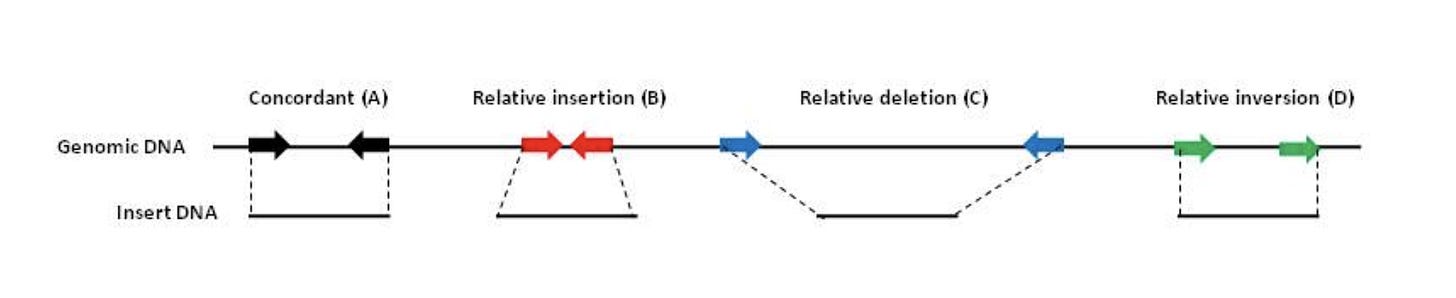
This is a field the team at Agencourt Personal Genomics pioneered as they were the first Next Generation sequencing platform (SOLiD sequencing) that used TypeIIs restriction enzymes to make paired end libraries that spanned 10-20kb in size. These large insert libraries could detect novel structural variations in human genomes (Figure 4).
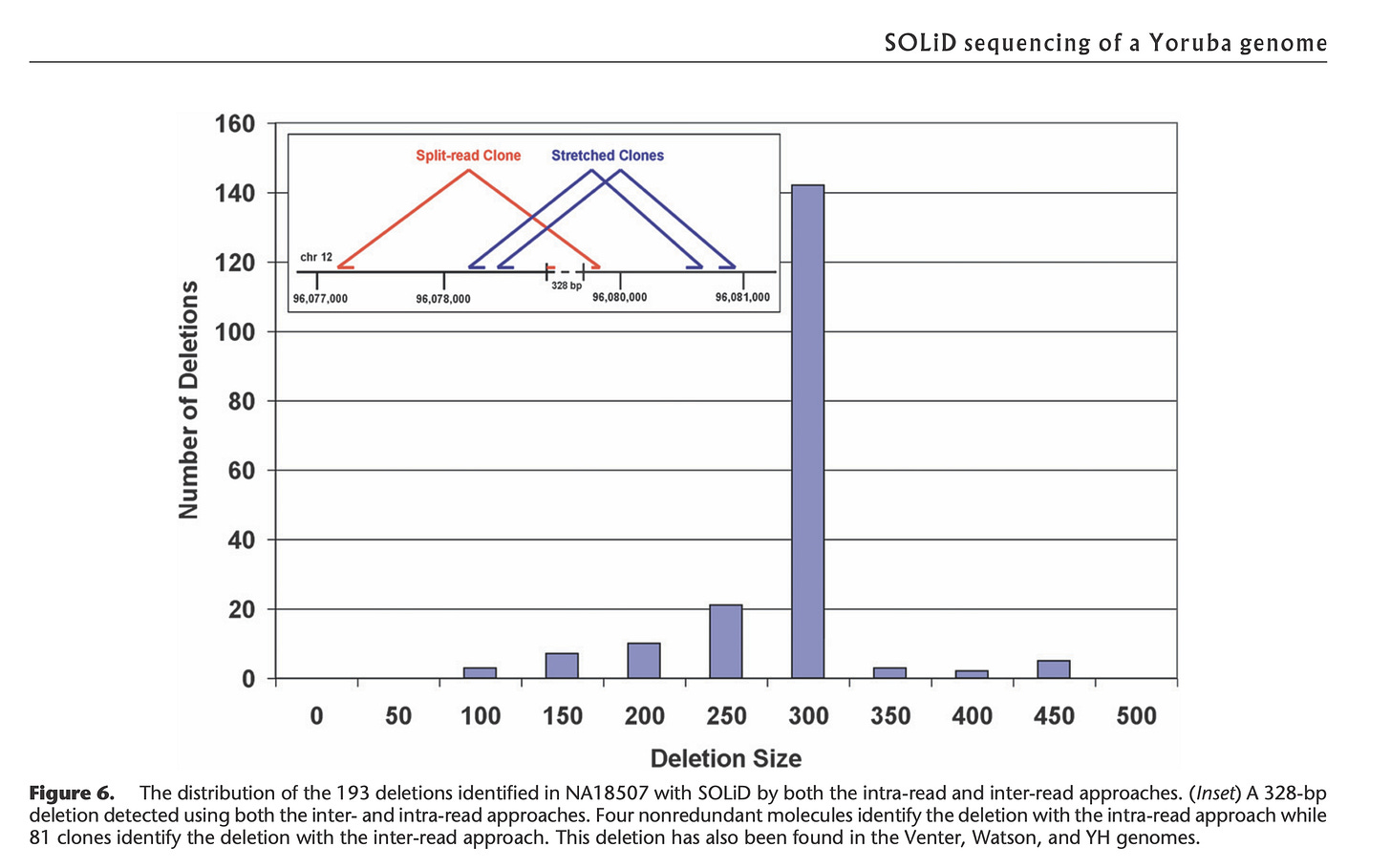
The Broad Institute (the authors of IGV:Integrated Genome Viewer used in Figure 1), have a great tutorial that covers read pairing with Illumina and SOLiD sequencing.
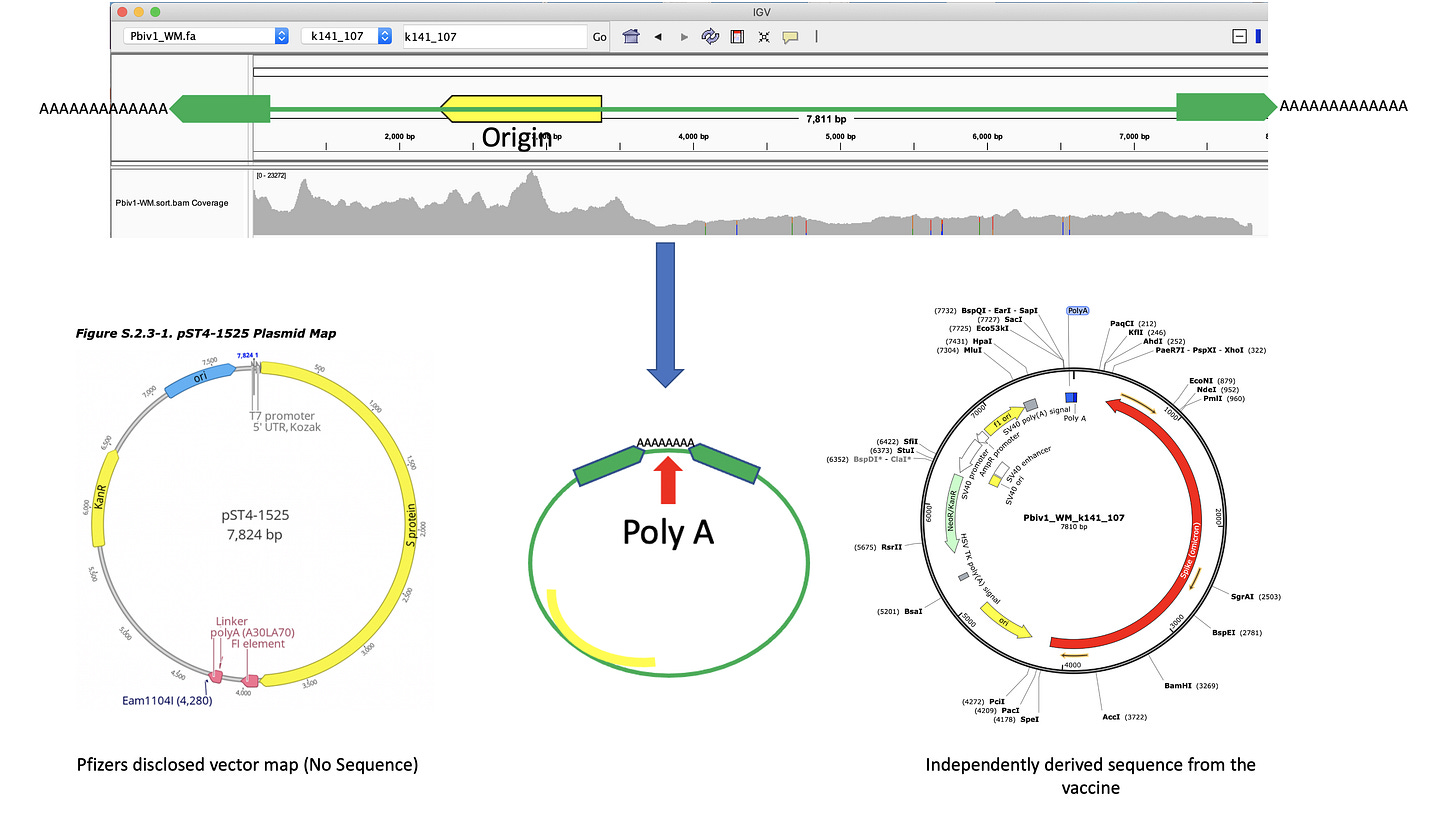
Figure 5 Illustrates Pfizer’s disclosed vector map (EMA documentation below) on the left. The vector on the right is what the RNase A sequencing and assembly produced. The middle plasmid is a depiction of the circularized read pairs from the Top track. The top track is a depiction of a linear plasmid assembly and the reads with in that assembly that are evidence of the circular state of the molecule. The fact that the green reads are pointing off the edges of the reference and we know they are 250bp molecules, leads to one structural solution as circular molecules. The origin of replication has higher coverage because these plasmids were isolated from E.coli in mid log scale growth (EMA document) and the assembly is capturing the fact that a population of the plasmids are in mid replication.
Background: The EMA
Many thanks to
for forwarding this EMA document. Note, on page 69, the regulators begin asking some pointy questions on the spotty genomics used to QC these products. The linearization reaction used to cut the plasmid with a restriction enzyme, is never quantitated.Other interesting details are disclosed regarding their process. We need a mechanism to monitor dsRNA as the Immunoassay methods they are using for this have very little literature on N1-methylpseudoU (m1Ψ) . If readers find any, please post it. Counter to the poorly described immunoassay in the EMA document, we have an RNA-seq strandedness analysis that demonstrates discrete regions in the mRNA that have more anti-sense alignments than sense alignments with the sequencing reads.
Several examples of concerns raised by the EMA that could lead to a failure to detect the high levels of dsDNA or dsRNA contamination in these vials.
This is an artifact of m1Ψ being a very sticky base. Note, replacing just 4 uracils with m1Ψ jumps the Tm nearly C. Change a 1000 and you have molecular velcro.
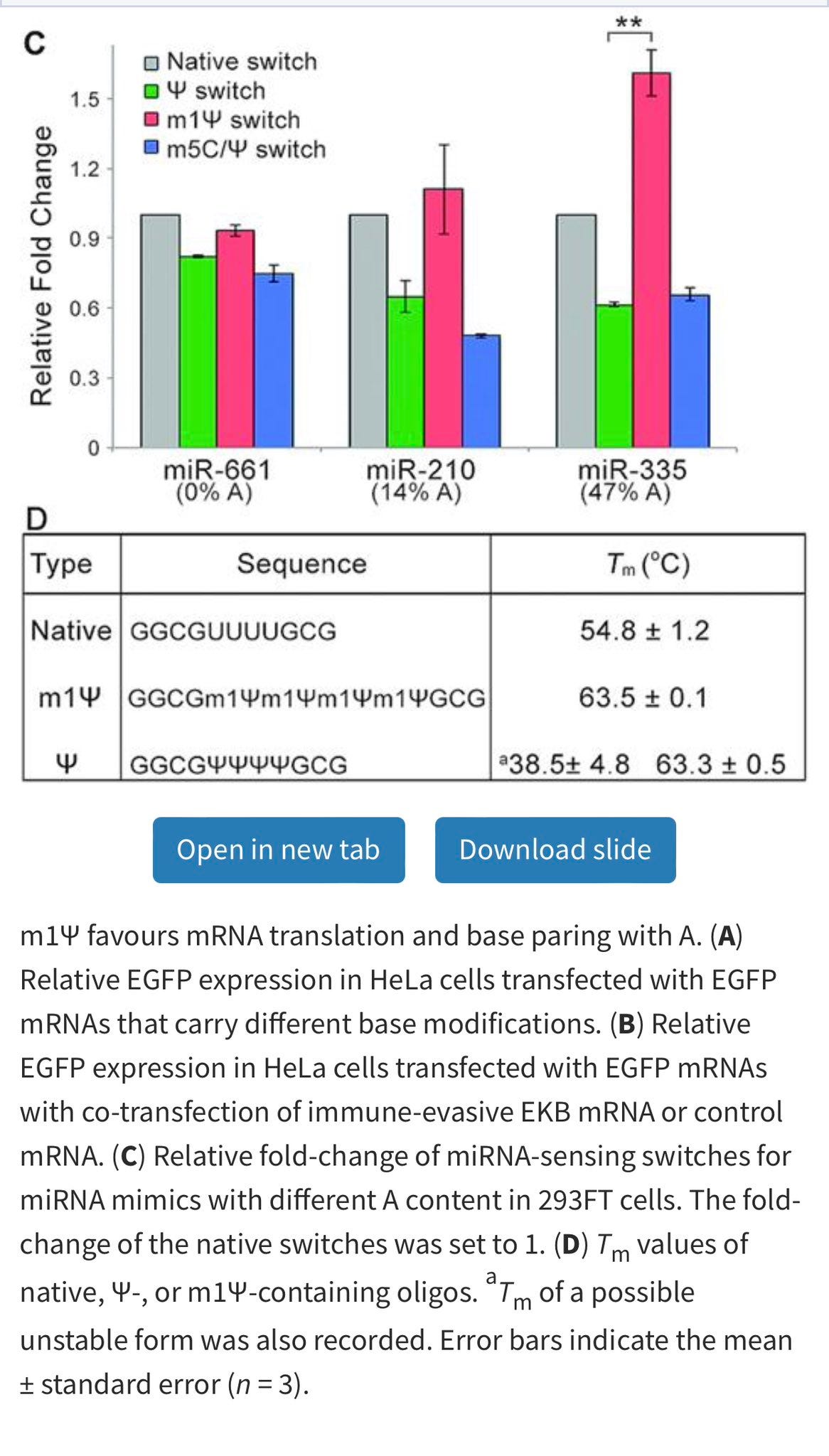
Pfizer’s sequencing QC work omits discussion of paired reads and if any reads bridge the cut site (Eam1104I). This cut site is adjacent to the Poly A tail and the above analysis demonstrates many reads which bridge this cut site. That can only happen on molecules that fail to cut and there are many present.
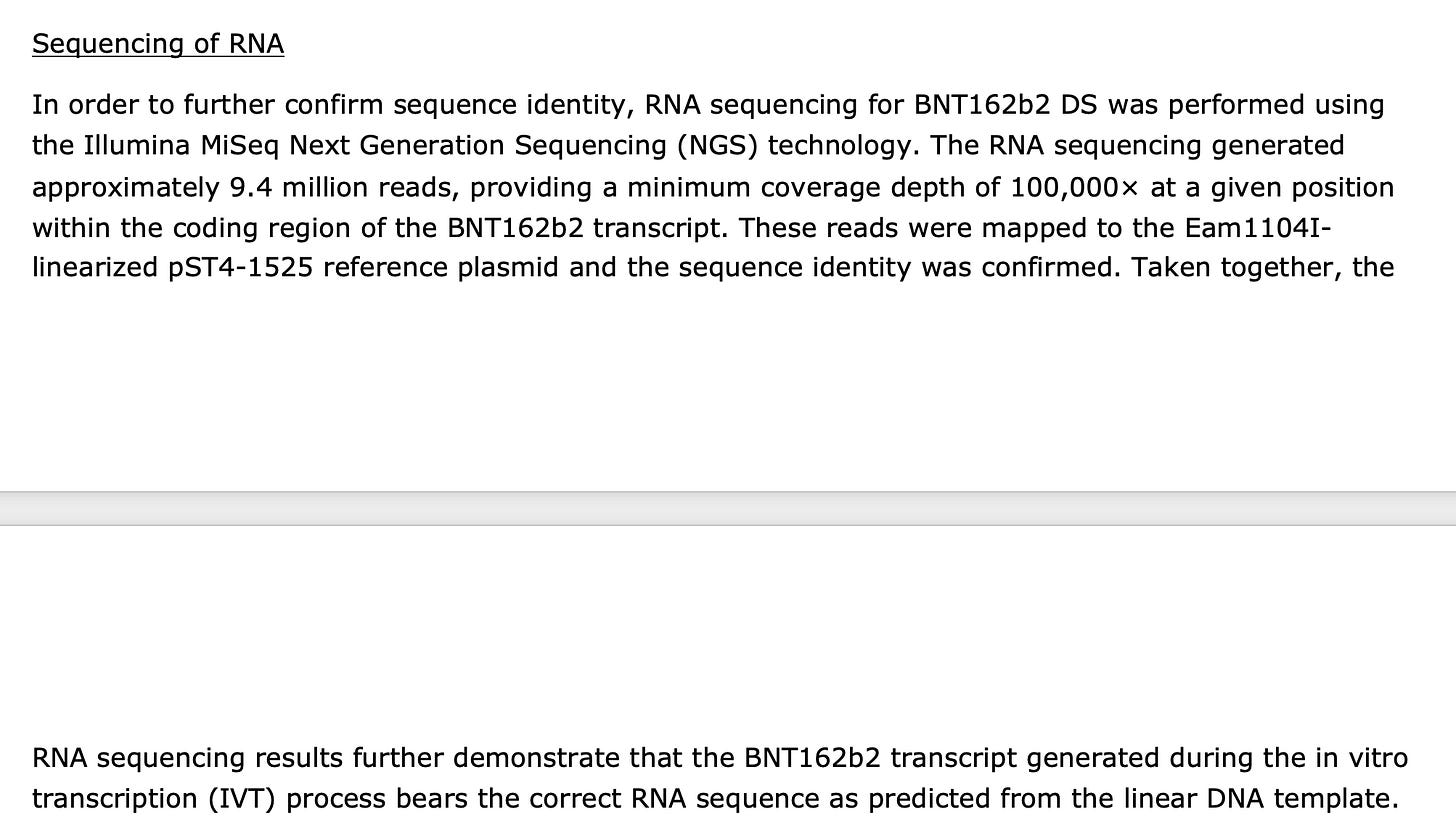
They claim to test for background DNA but are called out for providing no data nor any methods to back those claims.
This is also a classic magic trick that escapes many reviewers as they fail to appreciate that RT-qPCR amplifies both RNA and DNA. They are targeting spike with RT-qPCR. The spike sequence exists in both the plasmid vector and the spike mRNA. RT-qPCR amplifies both DNA and RNA so this assay will conflate the two.
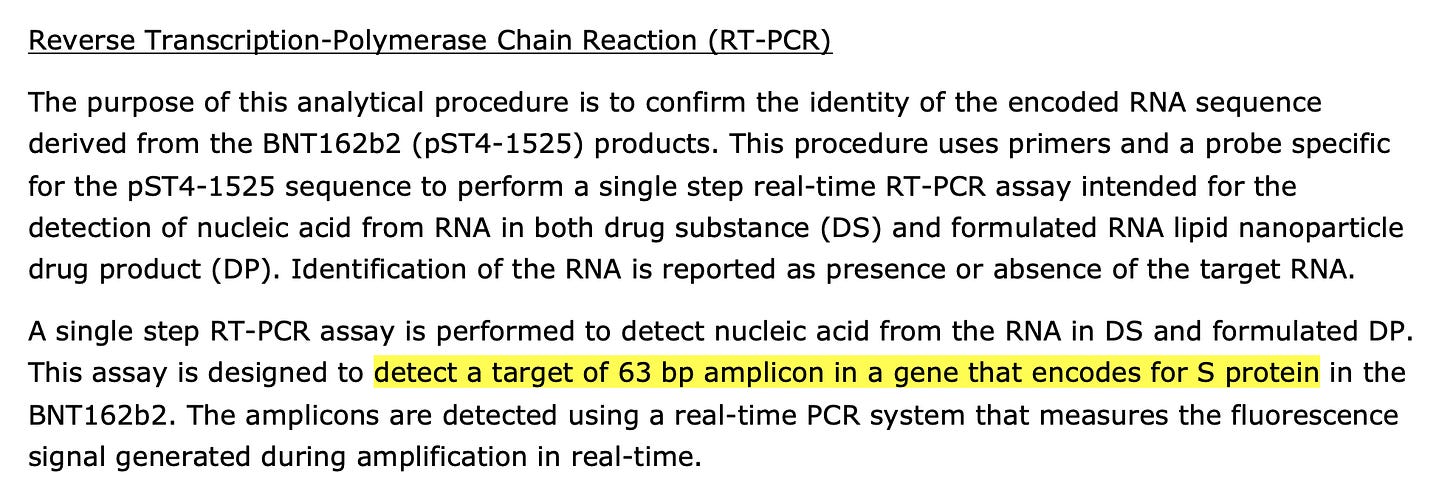
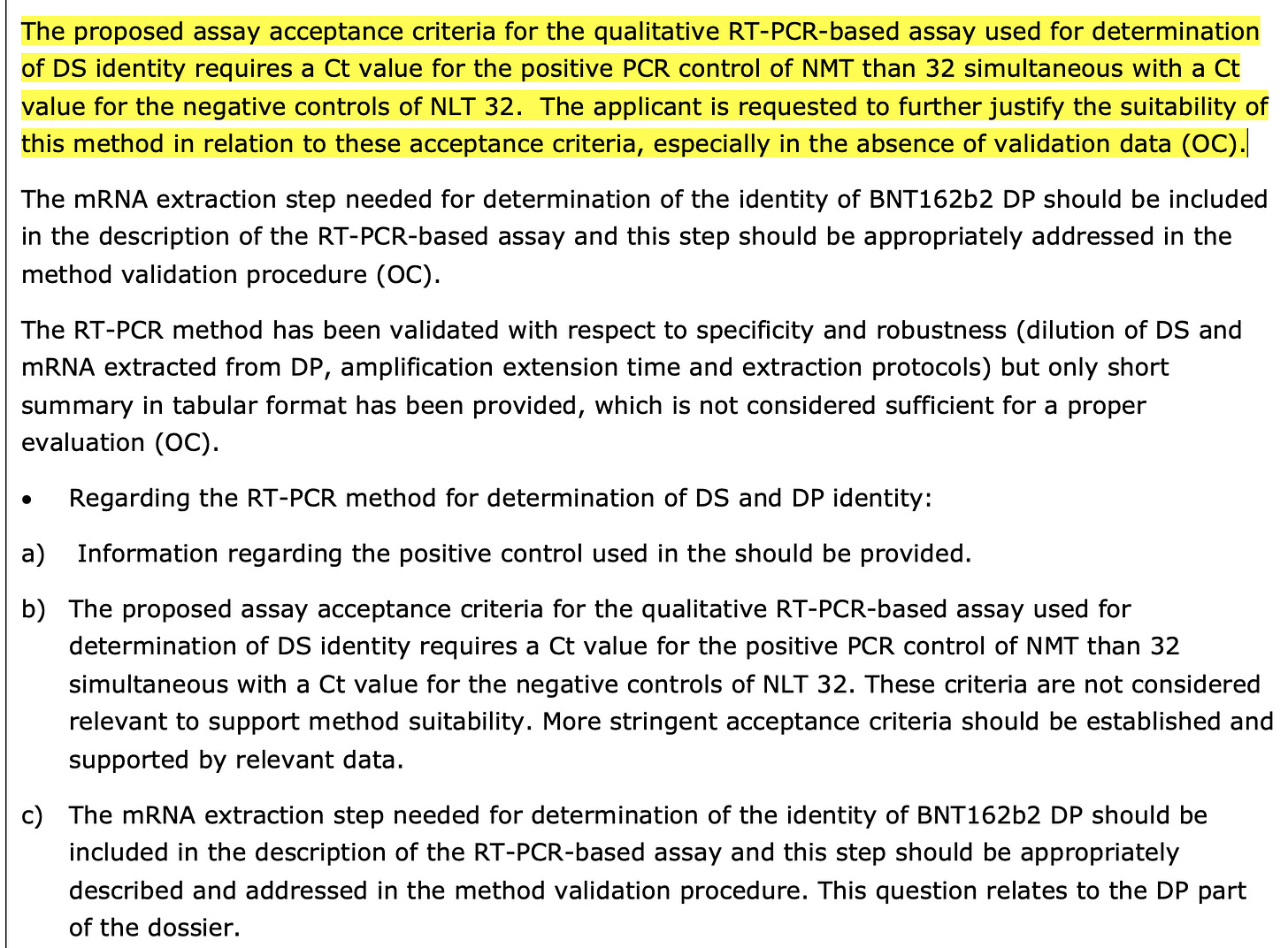
The regulators call them out for not disclosing the CT value. Notice they also don’t like CTs over 32 but don’t disclose the CT their samples produce.
The same goes for their qPCR looking at DNA. They don’t address linearity versus circles with this.
The residual dsDNA with these undisclosed methods also varies 10X.
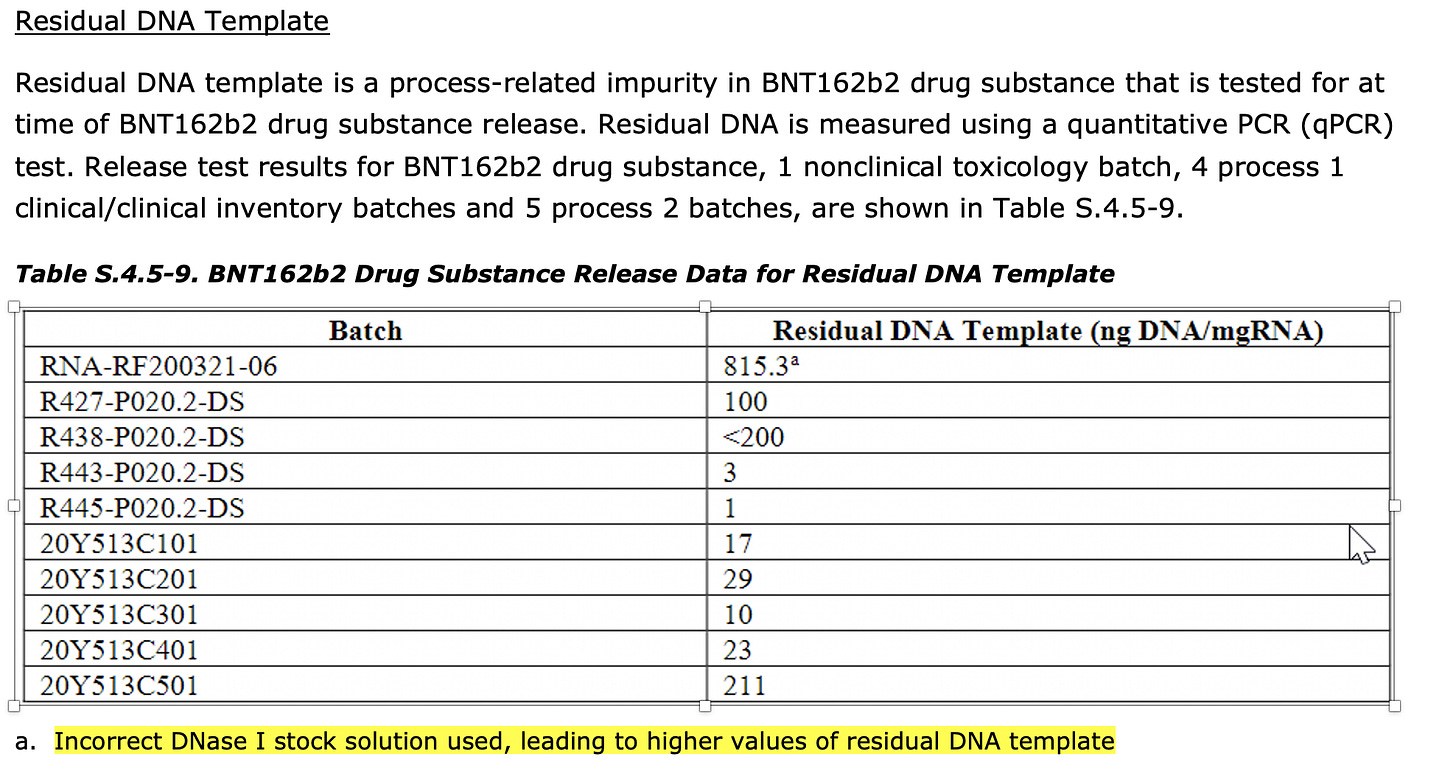
More evidence that their DNase I step is in question.
Results
With the mRNA removed from the sequencing, we now have a much cleaner picture of the vector sequence and must address a correction. Previous assemblies provided very short polyA tracts leading us to speculate that they must be adding the 100bp poly A on after expression from this plasmid. The RNase A libraries change this conclusion as long poly A tracts are in fact now in the vector. This is a PANDA presentation on the data before we had this information. Note, my hypothesis regarding their ligation of poly A tails on after T7 polymerization has been falsified with the RNase A libraries.


The Pfizer vaccine sequence is now 7810 bases. The Pfizer disclosure is just 14 bases longer (7824bp). The disagreement is in the linker in the Poly A tail. This is a 30A→10bp linker→ 70A region described by Nance et al. It is very likely Illumina sequencing exhibits polymerase slippage over this region and the disagreement is artifactual.

>k141_107 flag=1 multi=7745.0000 len=7810
TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCTAGCTCCAGGGTGTGGCTGGCACGAAATTGACCAACCCTGGGGTTAGTATAGCTTAGTTAAACTTTCGTTTATTGCTAAAGGTTAATCACTGCTGTTTCCCGTGGGGGTGTGGCTAGGCTAAGCGTTTTGAGCTGCATTGCTGCGTGCTTGGGAGGTGTCTGGAACTAGCAGAGGTGGTGAGTGGGGCAGGTGGAGGTGGGAGCATACCTGGGACCCGAGGTCGGGGGAGACTCGGGGTACCCAGGACGGGAAAGGGGCAGCTAGCATTGCGTGCATGCAGTACCAGCTCGAGTCATCATGTGTAGTGCAGTTTCACGCCCTTCAGCACGGGCTCAGAATCGTCCTCGTCGAACTTGCAGCAGCTGCCACAGCTACAACAGCCCTTCAGGCAGCTACAGCAGCTGGTCATGCAACACAGCATGATTGTGACCATCACGATGGCAATCAGTCCGGCGATAAAGCCCAGCCAGATGTACCAGGGCCACTTGATGTACTGCTCGTACTTCCCCAGTTCTTGCAGGTCGATCAGGCTCTCGTTCAGATTCTTGGCCACCTCGTTCAGCCGGTCGATCTCTTTCTGGATGTTCACGACGCTGGCATTGATTCCGCTGATATCGCCCAGGTCCACGTCGGGGCTTGTGTGGTTCTTAAAGTACTTGTCCAGTTCCTCTTTGAAGCTGTCCAGCTCGGGCTGCAGAGGGTCGTACACGGTATTGTTCACAATGCCGATCACGACGTCGCAGTTGCCAGACACGAAGGTGTTGTCGGTGGTGATGATCTGGGGCTCGTAGAAGTTCCGCTGTGTCACGAACCAATGGGTGCCGTTGGACACGAACACGCCTTCTCTAGGAAAGTGGGCTTTGCCGTCGTGGCAGATGGCTGGAGCGGTGGTGAAATTCTTCTCTTGAGCGGGCACATATGTCACGTGCAGAAACACCACGCCGTGAGGGGCAGACTGAGGGAAGCTCATCAGGTGGTAGCCCTTGCCGCAAAAGTCCACTCTCTTGCTCTGGCCCAGCACACACTCAGACATCTTGGTGGCGGCCAGATTGGCAGAGGCTCTAATCTCGGCGGCTCTGATCAGCTGCTGGGTCACGTATGTCTGGAGGCTCTGCAGTCTGCCTGTGATCAGTCTGTCGATCTGCACCTCGGCCTCAGGAGGGTCCAGTCTGCTCAGGATATCGTTCAGCACAGAGCTGATGGCGCCGAACTTGGAGGACAGCTGCTTGACCAGGGTGTTCAGTGCCTGGGCATTGTGGTTGACCACGTCCTGCAGCTTTCCCAGGGCGCTTGCTGTGCTGCTCAGGCTGTCCTGGATCTTGCCGATGGCGCTGTTGAACTGGTTGGCGATCAGCTTCTGGTTCTCGTACAGCACATTCTGGGTCACTCCGATGCCGTTGAACCGGTAGGCCATCTGCATAGCAAAGGGGATCTGCAGAGCGGCGCCTGCTCCAAATGTCCAGCCGCTTGTGATTGTGCCGGCCAGCAGGGCAGATGTGTACTGGGCGATCATCTCATCGGTCAGCAGAGGAGGCAGCACTGTCAGTCCGTTAAACTTCTGGGCGCAAATCAGATCCCTGGCGGCAATGTCGCCCAGACAATCGCCATACTGCTTGATGAAGCCGGCGTCGGCCAGTGTCACTTTGTTGAACAGCAGGTCCTCGATGAAGCTCCGCTTGCTGGGCTTGCTAGGATCGGGCAGAATCTGGCTGAAATTGAAGCCGCCGAAGTACTTGATAGGAGGGGTCTTGTAGATCTGCTTCACTTGGGCGAACACCTCTTGGGTGTTCTTGTCCTGTTCCACGGCGATCCCTGTCAGGGCTCTTTTCAGCTGGGTGCAGAAGCTGCCGTACTGCAGCAGCAGGTTGGAGCACTCGGTGGAATCGCCGCAGATGTACATGGTGCAGTCCACGCTGGTCTTGGTCATGGACACAGGCAGGATCTCTGTGGTCACGCTGATGGTGAAGTTGGTGGGGATAGCGATAGAGTTGTTGGAGTAGGCCACGCTGTTCTCGGCGCCCAGAGACATTGTGTAGGCAATGATGCTCTGGCTGGCCACGCTTCTGGCTCTCCGGTGGCTCTTTGTCTGTGTCTGGTAGCTGGCGCAGATTCCAGCGCCGATGGGGATGTCGCACTCGTAGCTATTGTTCACGTACTCGGCTCCGATCAGACAGCCGGCTCTGGTCTGAAACACATTGCTGCCGGTGGAGTACACCCGCCATGTAGGTGTCAGCTGATCGGCGTGAATGGCCACGGGCACTTCGGTACAGTTCACGCCCTGGTACAGCACTGCCACCTGATTGCTGGTGTTGGTGCCAGGGGTGATCACAGACACTCCGCCGAAGCTGCAAGGGGTGATGTCCAGGATTTCCAGTGTCTGGGGATCTCTAACGGCGTCTGTGGTATCGGCGATATCCCGGCCAAACTGCTGGAATGGCAGGAACTTCTTGTTGCTCTCTGTCAGCACGCCGGTGCCGGTCAGGCCGTTGAAGTTGAAGTTCACGCATTTGTTCTTCACGAGATTGGTGCTTTTCTTAGGGCCGCACACTGTGGCAGGGGCATGCAGCAGTTCGAAGCTCAGCACCACCACTCTGTAGGGCTGGTGGCCCACGCCGTATGTGGGCCTAAAGCCGTAGGACTGCAGTGGGAAGTAGCAGTTCACGCCTGCCACGCCGTTACAAGGCTTGTTGCCGGCCTGATAGATCTCGGTGGAGATGTCCCGCTCGAAGGGCTTCAGATTGGACTTCCGGAACAGCCGGTACCTGTAATTGTAGTTGCCGCCGACTTTGGAGTCCAGCTTGTTGCTGTTCCAGGCAATCACACAGCCGGTGAAGTCGTCGGGCAGCTTGTAGTTGTAGTCGGCGATGTTGCCTGTCTGTCCAGGGGCAATCTGCCGCACTTCGTTTCCCCGGATCACGAAGCTGTCGGCGTACACGTTTGTGAAGCACAGGTCGTTCAGCTTGGTAGGGGACACGCCGTAGCACTTGAATGCGAAGAAGGGGGCGAAGTTGTACAGCACGGAGTAGTCGGCCACGCAATTGCTGATCCGCTTCCGGTTCCAGGCGTACACAGAGGCGAATCTGGTGGCATTGAACACCTCGTCGAAGGGGCACAGATTGGTGATATTGGGGAACCGCACGATGGATTCGGTGGGCTGCACCCGGAAGTTGCTGGTCTGGTAGATGCCCTTTTCCACGGTGAAGGACTTCAGGGTGCACTTTGTCTCGCTCAGAGGATCCAGAGCACAATCCACGGCGTCGGTGATGGTGCCGTTCTCGTTGTACTTCAGCAGGAAGGTTCTAGGCTGCAGGTAGCCCACATAGTAAGCGGCGGCACCAGCTGTCCATCCGCTGCTGCTATCGCCAGGTGTCAGGTAGCTTCTGTGCAGGGCCAGCAGTGTCTGAAACCGGGTGATGTTGATGCCGATGGGCAGATCCACCAGGGGTTCCAGAGCAGAGAAGCCCTGAGGCAGATCCCGGCCGAGGTTGATAGGGGTGTGCTTGCTGTAGATCTTGAAGTAGCCGTCGATGTTCTTAAACACGAACTCGCGCAGGTTCTTGAAGTTGCCCTGCTTGCCTTCCAGGTCCATCAGGAAAGGCTGGGACACGTACTCGAAGGTGCAGTTGTTGGCGCTGCTGTACACCCGGAACTCGCTTTCCATCCAGCTCTTGTTGTTCTTGTGGTAGTAGACGTCCAGGAAGGGGTCGTTGCAGAACTGGAACTCGCACACTTTGATGACCACGTTGGTGGCGTTGTTCACGATCAGCAGGCTCTGGGTCTTGCTGTCCAGTGTGGTGCCGAAGATCCAGCCTCTGATGATGTTGGACTTCTCGGTGCTGGCAAAGTACACCCCGTCGTTGAAGGGCAGCACGGGGTTGTCGAATCTCTTGGTGCCATTGGTGCCGGAGATGGCGTGGAACCAGGTCACGTTGCTGAAGAAAGGCAGGAACAGGTCCTGGGTAGAGTGCAGCACGCTGGATCTGAACACCTTGTCGGGGTAGTACACGCCTCTGGTAAAGCTGTTGGTGTATGACTGTGTTCTGGTGATCAGGTTCACACACTGGCTGGACACCAGAGGCAGCAGCACCAGGAACACGAACATGGTGGCGGGTTCTCTCTGAGTCTGTGGGGACCAGAAGAATACTAGTTTATTCTTATAGTGAGTCGTATTAATTAATAACTAATGCATGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAACCTGAGGCTATGGCAGGGCCTGCCGCCCCGACGTTGGCTGCGAGCCCTGGGCCTTCACCCGAACTTGGGGGGTGGGGTGGGGAAAAGGAAGAAACGCGGGCGTATTGGCCCCAATGGGGTCTCGGTGGGGTATCGACAGAGTGCCAGCCCTGGGACCGAACCCCGCGTTTATGAACAAACGACCCAACACCGTGCGTTTTATTCTGTCTTTTTATTGCCGTCATAGCGCGGGTTCCTTCCGGTATTGTCTCCTTCCGTGTTTCAGTTAGCCTCCCCCTAGGGTGGGCGAAGAACTCCAGCATGAGATCCCCGCGCTGGAGGATCATCCAGCCGGCGTCCCGGAAAACGATTCCGAAGCCCAACCTTTCATAGAAGGCGGCGGTGGAATCGAAATCTCGTGATGGCAGGTTGGGCGTCGCTTGGTCGGTCATTTCGAACCCCAGAGTCCCGCTCAGAAGAACTCGTCAAGAAGGCGATAGAAGGCGATGCGCTGCGAATCGGGAGCGGCGATACCGTAAAGCACGAGGAAGCGGTCAGCCCATTCGCCGCCAAGTTCTTCAGCAATATCACGGGTAGCCAACGCTATGTCCTGATAGCGGTCCGCCACACCCAGCCGGCCACAGTCGATGAATCCAGAAAAGCGGCCATTTTCCACCATGATATTCGGCAAGCAGGCATCGCCATGGGTCACGACGAGATCCTCGCCGTCGGGCATGCTCGCCTTGAGCCTGGCGAACAGTTCGGCTGGCGCGAGCCCCTGATGTTCTTCGTCCAGATCATCCTGATCGACAAGACCGGCTTCCATCCGAGTACGTGCTCGCTCGATGCGATGTTTCGCTTGGTGGTCGAATGGGCAGGTAGCCGGATCAAGCGTATGCAGCCGCCGCATTGCATCAGCCATGATGGATACTTTCTCGGCAGGAGCAAGGTGAGATGACAGGAGATCCTGCCCCGGCACTTCGCCCAATAGCAGCCAGTCCCTTCCCGCTTCAGTGACAACGTCGAGCACAGCTGCGCAAGGAACGCCCGTCGTGGCCAGCCACGATAGCCGCGCTGCCTCGTCTTGCAGTTCATTCAGGGCACCGGACAGGTCGGTCTTGACAAAAAGAACCGGGCGCCCCTGCGCTGACAGCCGGAACACGGCGGCATCAGAGCAGCCGATTGTCTGTTGTGCCCAGTCATAGCCGAATAGCCTCTCCACCCAAGCGGCCGGAGAACCTGCGTGCAATCCATCTTGTTCAATCATGCGAAACGATCCTCATCCTGTCTCTTGATCGATCTTTGCAAAAGCCTAGGCCTCCAAAAAAGCCTCCTCACTACTTCTGGAATAGCTCAGAGGCCGAGGCGGCCTCGGCCTCTGCATAAATAAAAAAAATTAGTCAGCCATGGGGCGGAGAATGGGCGGAACTGGGCGGAGTTAGGGGCGGGATGGGCGGAGTTAGGGGCGGGACTATGGTTGCTGACTAATTGAGATGCATGCTTTGCATACTTCTGCCTGCTGGGGAGCCTGGGGACTTTCCACACCTGGTTGCTGACTAATTGAGATGCATGCTTTGCATACTTCTGCCTGCTGGGGAGCCTGGGGACTTTCCACACCCTAACTGACACACATTCCACAGCTGGTTCTTTCCGCCTCAGGATTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTGTGGTGGTTACGCGCAGCGTGACCGCTACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTTAGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGTGATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAAGGGATTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAATTTAACGCGAATTTTAACAAAATATTAACGCTTACAATTTACGCGTTAAGATACATTGATGAGTTTGGACAAACCACAACTAGAATGCAGTGAAAAAAATGCTTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTAACCATTATAAGCTGCAATAAACAAGTTAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCAGGGGGAGGTGTGGGAGGTTTTTTAAAGCAAGTAAAACCTCTACAAATGTGGTATGGCTGATTATGATCATGAACAGACTGTGAGGACTGAGGGGCCTGAAATGAGCCTTGGGACTGTGAATCTAAAATACACAAACAATTAGAATCAGTAGTTTAACACATTATACACTTAAAAATTGGATCTCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATGTTAAACGGCGGAGCTACCACACCGGTTGGAGCTCTTCTTTTTTTTTTTTTTTTTTTTGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT
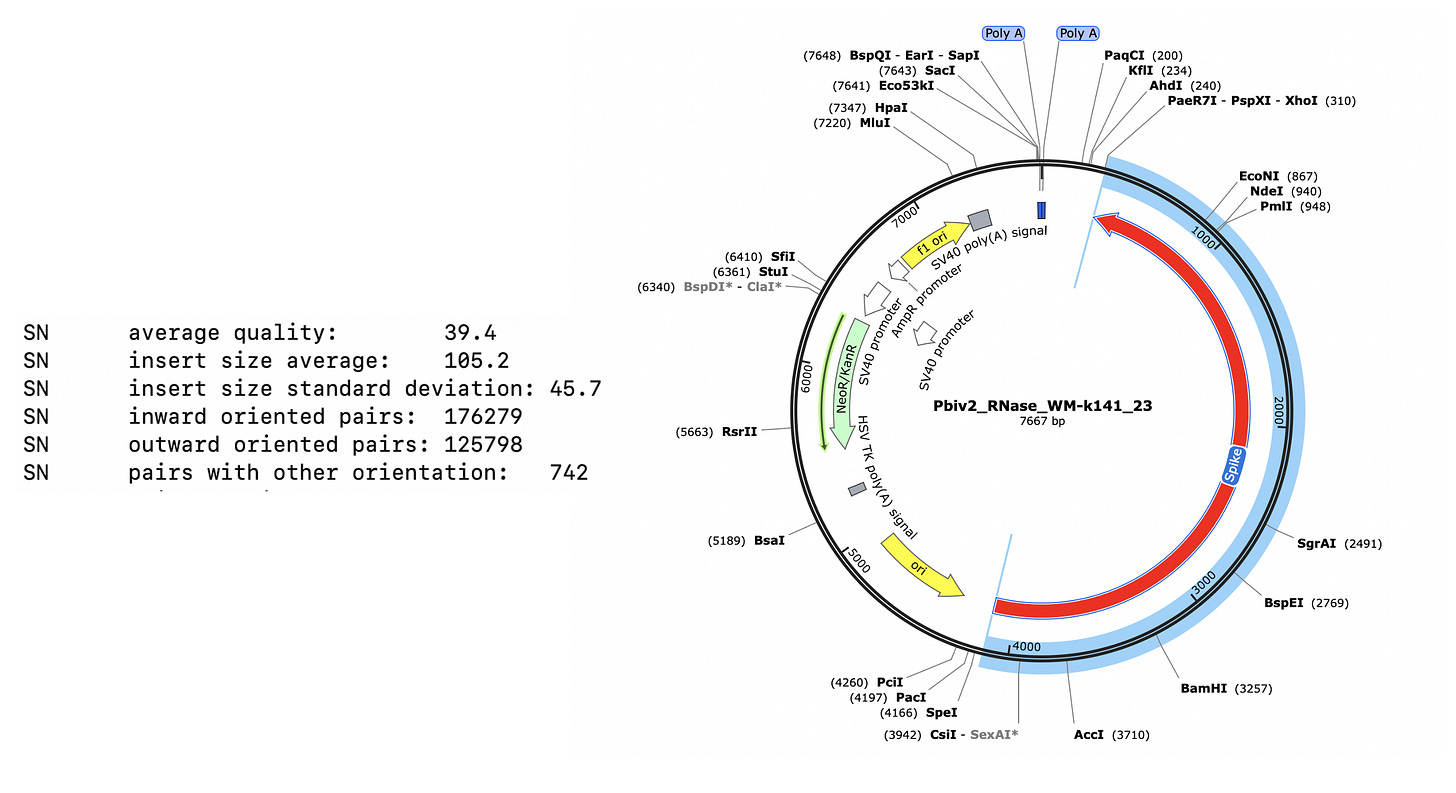
>k141_23 flag=1 multi=6914.0000 len=7667
TTTTTTTTTTTTTTTTTTTTGCTAGCTCCAGGGTGTGGCTGGCACGAAATTGACCAACCCTGGGGTTAGTATAGCTTAGTTAAACTTTCGTTTATTGCTAAAGGTTAATCACTGCTGTTTCCCGTGGGGGTGTGGCTAGGCTAAGCGTTTTGAGCTGCATTGCTGCGTGCTTGGGAGGTGTCTGGAACTAGCAGAGGTGGTGAGTGGGGCAGGTGGAGGTGGGAGCATACCTGGGACCCGAGGTCGGGGGAGACTCGGGGTACCCAGGACGGGAAAGGGGCAGCTAGCATTGCGTGCATGCAGTACCAGCTCGAGTCATCATGTGTAGTGCAGTTTCACGCCCTTCAGCACGGGCTCAGAATCGTCCTCGTCGAACTTGCAGCAGCTGCCACAGCTACAACAGCCCTTCAGGCAGCTACAGCAGCTGGTCATGCAACACAGCATGATTGTGACCATCACGATGGCAATCAGTCCGGCGATAAAGCCCAGCCAGATGTACCAGGGCCACTTGATGTACTGCTCGTACTTCCCCAGTTCTTGCAGGTCGATCAGGCTCTCGTTCAGATTCTTGGCCACCTCGTTCAGCCGGTCGATCTCTTTCTGGATGTTCACGACGCTGGCATTGATTCCGCTGATATCGCCCAGGTCCACGTCGGGGCTTGTGTGGTTCTTAAAGTACTTGTCCAGTTCCTCTTTGAAGCTGTCCAGCTCGGGCTGCAGAGGGTCGTACACGGTATTGTTCACAATGCCGATCACGACGTCGCAGTTGCCAGACACGAAGGTGTTGTCGGTGGTGATGATCTGGGGCTCGTAGAAGTTCCGCTGTGTCACGAACCAATGGGTGCCGTTGGACACGAACACGCCTTCTCTAGGAAAGTGGGCTTTGCCGTCGTGGCAGATGGCTGGAGCGGTGGTGAAATTCTTCTCTTGAGCGGGCACATATGTCACGTGCAGAAACACCACGCCGTGAGGGGCAGACTGAGGGAAGCTCATCAGGTGGTAGCCCTTGCCGCAAAAGTCCACTCTCTTGCTCTGGCCCAGCACACACTCAGACATCTTGGTGGCGGCCAGATTGGCAGAGGCTCTAATCTCGGCGGCTCTGATCAGCTGCTGGGTCACGTATGTCTGGAGGCTCTGCAGTCTGCCTGTGATCAGTCTGTCGATCTGCACCTCGGCCTCAGGAGGGTCCAGTCTGCTCAGGATATCGTTCAGCACAGAGCTGATGGCGCCGAACTTGGAGGACAGCTGCTTGACCAGGGTGTTCAGTGCCTGGGCATTGTGGTTGACCACGTCCTGCAGCTTTCCCAGGGCGCTTGCTGTGCTGCTCAGGCTGTCCTGGATCTTGCCGATGGCGCTGTTGAACTGGTTGGCGATCAGCTTCTGGTTCTCGTACAGCACATTCTGGGTCACTCCGATGCCGTTGAACCGGTAGGCCATCTGCATAGCAAAGGGGATCTGCAGAGCGGCGCCTGCTCCAAATGTCCAGCCGCTTGTGATTGTGCCGGCCAGCAGGGCAGATGTGTACTGGGCGATCATCTCATCGGTCAGCAGAGGAGGCAGCACTGTCAGTCCGTTAAACTTCTGGGCGCAAATCAGATCCCTGGCGGCAATGTCGCCCAGACAATCGCCATACTGCTTGATGAAGCCGGCGTCGGCCAGTGTCACTTTGTTGAACAGCAGGTCCTCGATGAAGCTCCGCTTGCTGGGCTTGCTAGGATCGGGCAGAATCTGGCTGAAATTGAAGCCGCCGAAGTACTTGATAGGAGGGGTCTTGTAGATCTGCTTCACTTGGGCGAACACCTCTTGGGTGTTCTTGTCCTGTTCCACGGCGATCCCTGTCAGGGCTCTTTTCAGCTGGGTGCAGAAGCTGCCGTACTGCAGCAGCAGGTTGGAGCACTCGGTGGAATCGCCGCAGATGTACATGGTGCAGTCCACGCTGGTCTTGGTCATGGACACAGGCAGGATCTCTGTGGTCACGCTGATGGTGAAGTTGGTGGGGATAGCGATAGAGTTGTTGGAGTAGGCCACGCTGTTCTCGGCGCCCAGAGACATTGTGTAGGCAATGATGCTCTGGCTGGCCACGCTTCTGGCTCTCCGGTGGCTCTTTGTCTGTGTCTGGTAGCTGGCGCAGATTCCAGCGCCGATGGGGATGTCGCACTCGTAGCTATTGTTCACGTACTCGGCTCCGATCAGACAGCCGGCTCTGGTCTGAAACACATTGCTGCCGGTGGAGTACACCCGCCATGTAGGTGTCAGCTGATCGGCGTGAATGGCCACGGGCACTTCGGTACAGTTCACGCCCTGGTACAGCACTGCCACCTGATTGCTGGTGTTGGTGCCAGGGGTGATCACAGACACTCCGCCGAAGCTGCAAGGGGTGATGTCCAGGATTTCCAGTGTCTGGGGATCTCTAACGGCGTCTGTGGTATCGGCGATATCCCGGCCAAACTGCTGGAATGGCAGGAACTTCTTGTTGCTCTCTGTCAGCACGCCGGTGCCGGTCAGGCCGTTGAAGTTGAAGTTCACGCATTTGTTCTTCACGAGATTGGTGCTTTTCTTAGGGCCGCACACTGTGGCAGGGGCATGCAGCAGTTCGAAGCTCAGCACCACCACTCTGTAGGGCTGGTGGCCCACGCCGTATGTGGGCCTAAAGCCGTAGGACTGCAGTGGGAAGTAGCAGTTCACGCCTGCCACGCCGTTACAAGGCTTGTTGCCGGCCTGATAGATCTCGGTGGAGATGTCCCGCTCGAAGGGCTTCAGATTGGACTTCCGGAACAGCCGGTACCTGTAATTGTAGTTGCCGCCGACTTTGGAGTCCAGCTTGTTGCTGTTCCAGGCAATCACACAGCCGGTGAAGTCGTCGGGCAGCTTGTAGTTGTAGTCGGCGATGTTGCCTGTCTGTCCAGGGGCAATCTGCCGCACTTCGTTTCCCCGGATCACGAAGCTGTCGGCGTACACGTTTGTGAAGCACAGGTCGTTCAGCTTGGTAGGGGACACGCCGTAGCACTTGAATGCGAAGAAGGGGGCGAAGTTGTACAGCACGGAGTAGTCGGCCACGCAATTGCTGATCCGCTTCCGGTTCCAGGCGTACACAGAGGCGAATCTGGTGGCATTGAACACCTCGTCGAAGGGGCACAGATTGGTGATATTGGGGAACCGCACGATGGATTCGGTGGGCTGCACCCGGAAGTTGCTGGTCTGGTAGATGCCCTTTTCCACGGTGAAGGACTTCAGGGTGCACTTTGTCTCGCTCAGAGGATCCAGAGCACAATCCACGGCGTCGGTGATGGTGCCGTTCTCGTTGTACTTCAGCAGGAAGGTTCTAGGCTGCAGGTAGCCCACATAGTAAGCGGCGGCACCAGCTGTCCATCCGCTGCTGCTATCGCCAGGTGTCAGGTAGCTTCTGTGCAGGGCCAGCAGTGTCTGAAACCGGGTGATGTTGATGCCGATGGGCAGATCCACCAGGGGTTCCAGAGCAGAGAAGCCCTGAGGCAGATCCCGGCCGAGGTTGATAGGGGTGTGCTTGCTGTAGATCTTGAAGTAGCCGTCGATGTTCTTAAACACGAACTCGCGCAGGTTCTTGAAGTTGCCCTGCTTGCCTTCCAGGTCCATCAGGAAAGGCTGGGACACGTACTCGAAGGTGCAGTTGTTGGCGCTGCTGTACACCCGGAACTCGCTTTCCATCCAGCTCTTGTTGTTCTTGTGGTAGTAGACGTCCAGGAAGGGGTCGTTGCAGAACTGGAACTCGCACACTTTGATGACCACGTTGGTGGCGTTGTTCACGATCAGCAGGCTCTGGGTCTTGCTGTCCAGTGTGGTGCCGAAGATCCAGCCTCTGATGATGTTGGACTTCTCGGTGCTGGCAAAGTACACCCCGTCGTTGAAGGGCAGCACGGGGTTGTCGAATCTCTTGGTGCCATTGGTGCCGGAGATGGCGTGGAACCAGGTCACGTTGCTGAAGAAAGGCAGGAACAGGTCCTGGGTAGAGTGCAGCACGCTGGATCTGAACACCTTGTCGGGGTAGTACACGCCTCTGGTAAAGCTGTTGGTGTATGACTGTGTTCTGGTGATCAGGTTCACACACTGGCTGGACACCAGAGGCAGCAGCACCAGGAACACGAACATGGTGGCGGGTTCTCTCTGAGTCTGTGGGGACCAGAAGAATACTAGTTTATTCTTATAGTGAGTCGTATTAATTAATAACTAATGCATGGCGGTAATACGGTTATCCACAGAATCAGGGGATAACGCAGGAAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGTTTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCCCCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCATAGCTCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTAACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTACAGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGTAGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTTTGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGAGATTATCAAAAAGGATCTTCACCTAGATCCTTTTAAATTAAAAATGAAGTTTTAAATCAATCTAAAGTATATATGAGTAACCTGAGGCTATGGCAGGGCCTGCCGCCCCGACGTTGGCTGCGAGCCCTGGGCCTTCACCCGAACTTGGGGGGTGGGGTGGGGAAAAGGAAGAAACGCGGGCGTATTGGCCCCAATGGGGTCTCGGTGGGGTATCGACAGAGTGCCAGCCCTGGGACCGAACCCCGCGTTTATGAACAAACGACCCAACACCGTGCGTTTTATTCTGTCTTTTTATTGCCGTCATAGCGCGGGTTCCTTCCGGTATTGTCTCCTTCCGTGTTTCAGTTAGCCTCCCCCTAGGGTGGGCGAAGAACTCCAGCATGAGATCCCCGCGCTGGAGGATCATCCAGCCGGCGTCCCGGAAAACGATTCCGAAGCCCAACCTTTCATAGAAGGCGGCGGTGGAATCGAAATCTCGTGATGGCAGGTTGGGCGTCGCTTGGTCGGTCATTTCGAACCCCAGAGTCCCGCTCAGAAGAACTCGTCAAGAAGGCGATAGAAGGCGATGCGCTGCGAATCGGGAGCGGCGATACCGTAAAGCACGAGGAAGCGGTCAGCCCATTCGCCGCCAAGTTCTTCAGCAATATCACGGGTAGCCAACGCTATGTCCTGATAGCGGTCCGCCACACCCAGCCGGCCACAGTCGATGAATCCAGAAAAGCGGCCATTTTCCACCATGATATTCGGCAAGCAGGCATCGCCATGGGTCACGACGAGATCCTCGCCGTCGGGCATGCTCGCCTTGAGCCTGGCGAACAGTTCGGCTGGCGCGAGCCCCTGATGTTCTTCGTCCAGATCATCCTGATCGACAAGACCGGCTTCCATCCGAGTACGTGCTCGCTCGATGCGATGTTTCGCTTGGTGGTCGAATGGGCAGGTAGCCGGATCAAGCGTATGCAGCCGCCGCATTGCATCAGCCATGATGGATACTTTCTCGGCAGGAGCAAGGTGAGATGACAGGAGATCCTGCCCCGGCACTTCGCCCAATAGCAGCCAGTCCCTTCCCGCTTCAGTGACAACGTCGAGCACAGCTGCGCAAGGAACGCCCGTCGTGGCCAGCCACGATAGCCGCGCTGCCTCGTCTTGCAGTTCATTCAGGGCACCGGACAGGTCGGTCTTGACAAAAAGAACCGGGCGCCCCTGCGCTGACAGCCGGAACACGGCGGCATCAGAGCAGCCGATTGTCTGTTGTGCCCAGTCATAGCCGAATAGCCTCTCCACCCAAGCGGCCGGAGAACCTGCGTGCAATCCATCTTGTTCAATCATGCGAAACGATCCTCATCCTGTCTCTTGATCGATCTTTGCAAAAGCCTAGGCCTCCAAAAAAGCCTCCTCACTACTTCTGGAATAGCTCAGAGGCCGAGGCGGCCTCGGCCTCTGCATAAATAAAAAAAATTAGTCAGCCATGGGGCGGAGAATGGGCGGAACTGGGCGGAGTTAGGGGCGGGATGGGCGGAGTTAGGGGCGGGACTATGGTTGCTGACTAATTGAGATGCATGCTTTGCATACTTCTGCCTGCTGGGGAGCCTGGGGACTTTCCACACCCTAACTGACACACATTCCACAGCTGGTTCTTTCCGCCTCAGGATTCTTCCTTTTTCAATATTATTGAAGCATTTATCAGGGTTATTGTCTCATGAGCGGATACATATTTGAATGTATTTAGAAAAATAAACAAATAGGGGTTCCGCGCACATTTCCCCGAAAAGTGCCACCTGACGCGCCCTGTAGCGGCGCATTAAGCGCGGCGGGTGTGGTGGTTACGCGCAGCGTGACCGCTACACTTGCCAGCGCCCTAGCGCCCGCTCCTTTCGCTTTCTTCCCTTCCTTTCTCGCCACGTTCGCCGGCTTTCCCCGTCAAGCTCTAAATCGGGGGCTCCCTTTAGGGTTCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGTGATGGTTCACGTAGTGGGCCATCGCCCTGATAGACGGTTTTTCGCCCTTTGACGTTGGAGTCCACGTTCTTTAATAGTGGACTCTTGTTCCAAACTGGAACAACACTCAACCCTATCTCGGTCTATTCTTTTGATTTATAAGGGATTTTGCCGATTTCGGCCTATTGGTTAAAAAATGAGCTGATTTAACAAAAATTTAACGCGAATTTTAACAAAATATTAACGCTTACAATTTACGCGTTAAGATACATTGATGAGTTTGGACAAACCACAACTAGAATGCAGTGAAAAAAATGCTTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTAACCATTATAAGCTGCAATAAACAAGTTAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCAGGGGGAGGTGTGGGAGGTTTTTTAAAGCAAGTAAAACCTCTACAAATGTGGTATGGCTGATTATGATCATGAACAGACTGTGAGGACTGAGGGGCCTGAAATGAGCCTTGGGACTGTGAATCTAAAATACACAAACAATTAGAATCAGTAGTTTAACACATTATACACTTAAAAATTGGATCTCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATGTTAAACGGCGGAGCTACCACACCGGTTGGAGCTCTTCTTTTTTTTTTTTTTTTTTTT
These two consensus sequences differ slightly in their length of the poly A tail but most strikingly with a tandem duplication in the SV40 promoter of Vial 1 compared to Vial 2.
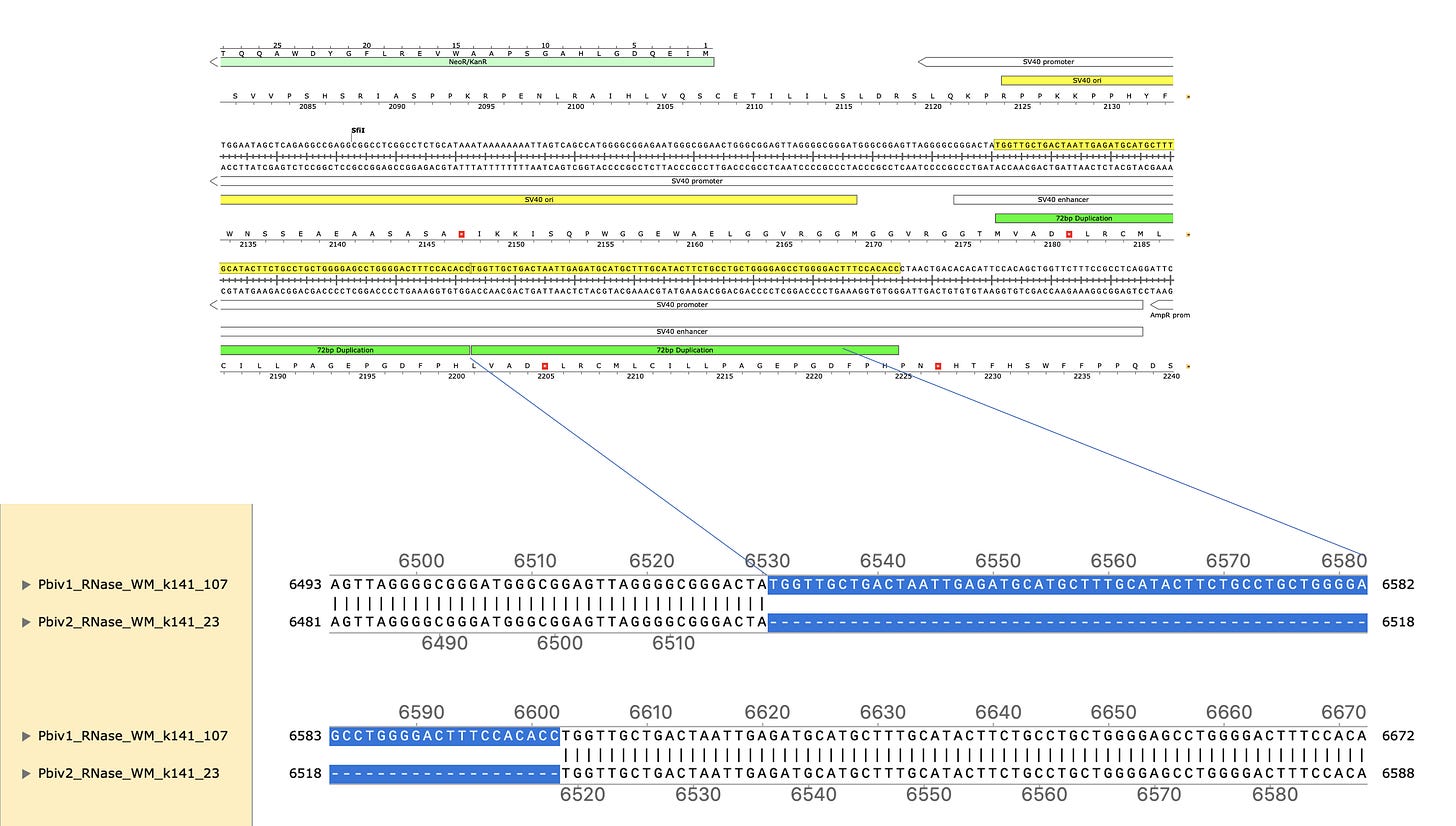
This is a well known way to turbo SV40. Sampling just two Pfizer vials delivers two different expression vectors? These vials are from the same lot. The inserts are 100% identical and unrelated to bivalent vaccines. I wonder how many more plasmids are out there?
The 72bp repeat is known to facilitate nuclear import.
Conclusions
1)Paired end reads support the existence of circular plasmids in the Pfizer bivalent vaccines. This is supported by transformation in kanamycin sensitive E.coli. The exact ratio of circular versus linear DNA is still being evaluated but the levels of DNA contamination exceed the EMA specifications by several orders of magnitude.
2)EMA documentation reveals regulators are unsatisfied with the molecular methods used to evaluate linearization. There are concerns over non specific CT values (<32) in the PCR methods used for quantification. Additional concerns are voiced over the variation in DNase I activity and methods used to evaluate its performance.
3)A long poly A tract is evident in the vectors reducing the odds of off target poly adenylation. The 10bp linker isn’t well resolved with Illumina sequencing and the two vectors also differ in this artifactual regard.
4)A known SV40 promoter improvement is evident in 1/2 of the vials from the same lot of vaccine.
Limitations.
While RNase A is effective at removing RNA, there are conditions in which it can also eliminate DNA. This may be pertinent to how Pfizer is assessing their dsDNA contamination as they are using a single RNase and the literature on RNase A and m1Ψ hybrids with DNA is sparse. More controls with alternative RNases like RNase R will help refine the estimates of circular to linear DNA.
Methods
Vaccine purification, RNase A treatment and sequencing methods were published previously but is repeated here for clarity.
RNase A treatment of the Vaccines
RNase A cleaves both uracils and cytosines. N1-methylpseudouridine is known to be RNAse-L resistant but RNase A will cleave cytosines which still exist in the mRNAs. This leaves predominantly DNA for sequencing. Vaccine mRNA that was previously sequenced and discussed here, was treated at 37C for 20 minutes with 3ul of 20 Units/ul Monarch RNase A from NEB. The RNase reaction was purified using 1.5X of SenSATIVAx (Medicinal Genomics #420001). Sample were eluted in 20ul ddH20 after DNA purification. 15ul was used for DNA sequencing and 5ul used for transformation of E.coli.
Whole genome shotgun of Pre-transformation RNase’d Vaccines.
15ul of the DNA (prior to E.coli transformation) was converted into sequence ready libraries using Watchmakers Genomics WGS library construction kit.
Analysis pipeline
Reads were demultiplexed and processed with
Trimgalore - Removes Illumina Sequencing adaptors.
Megahit- assembles reads into contigs.
Samtools- generates BAM files for viewing in IGV.
Samtools stats used to calculate outie reads.
BWA-mem- Short read mapper used to align reads back to the assembled references.
SnapGene software- (www.snapgene.com)- Used to visualize and annotate expression vectors
IGV- Integrated Genome Viewer used to visualize Illumina sequencing reads.
BAM files
contig specific BAM files were created using samtools
samtools view -h input.bam contig_name -O BAM > contig.bam; samtools index contig.bam;Samtools stats run on a each contig in each assembly.
for out_prefix in `ls *.sort.bam | perl -pe "s/.sort.bam//"`; do mkdir -p ${out_prefix}-samtools-stats; for contig in `samtools view -H ${out_prefix}.sort.bam | grep "^@SQ" | cut -f 2 | perl -pe "s/SN\://"`; do echo "Now calculating stats for ${contig}/$out_prefix..."; samtools stats ${out_prefix}.sort.bam $contig > ${out_prefix}-samtools-stats/${contig}-samtools-stats.txt; done; doneThe links above were destroyed by the New Zealand Ministry of Health, in their reckless attempt to destroy Steve Kirsch’s transformed vaccine death data. This destruction occurred prior to any injunction being issued and it appears the injunction was illegal and may expose the hosting company and the ERA/NZMH to discovery.
These data have been mirrored onto IPFS.
Posts script- Vial 2 appears to be heteroplasmic for the 72bp insertion. Note. Vial 1 is depicted in the opposite orientation as Vial 1.
Jalview for viewing the annotated vector sequences.





Anandamide, you says that the plasmid contamination is a proxy for LPS contamination.
As a dinosaur in age, I remember back to the 60's when the NCI scientists were making L-asparaginase to be used against cancer, primarily leukemia, lymphomas, melanomas, breast cancer etc...
Except it was causing huge problems for patients, including the liver, kidneys, blood clotting system and the brain, and central nervous system issues.... There were a lot of sudden deaths....
A bright spark suggested that it was E.coli LPS toxicity to which they laughed and said that "the amounts wouldn't hurt a monkey" Dr Gordon Zubrod was in charge...
Anyway, Zubrod wouldn't listen, until three years later Dr Tibor Borsos proved that 13 of 15 commercial lots of L-Asparaginase were contaminated with E.Coli Endotoxin.
all that time, they were still using it, and killing patients with it. All during that time, none of the oncologists could see the LPS toxic syndrome for it punching them on the nose.
At some point the light-bulb lit, and they changed the manufacturing process and used Erwinia Carotovora instead, and suddenly all the "mysterious" toxic deaths stopped.
Another story buried in the "no fishing" cupboard to remind us just what can happen when people don't appear to understand or take note of - or perhaps just ignore.... the basics of E.coli LPS toxicity.
Well done, again! Not surprising you have confirmed the presence of circular plasmid by sequencing given your earlier results, but it did need to be done. Also nice to have an improved assembly of their vector sequence and solve the polyA "mystery".
A stand out for me is still the quantity of DNA you detected in your previous Substack. Circular or linear, I still think this will happily transfect human cells, in some cases permanently altering the genome.
I was left wondering how the heck it could be that there is that much DNA left there, and also what kind of QC parameters they have for residual DNA in the first place. After perusing some Australian TGA FOI documents yesterday, and quickly reading through the EMA document you've linked here, this is basic summary as I see it:
1) The manufacturing process uses a linearised DNA plasmid together with T7 polymerase and other reagents to generate the RNA.
2) The removal of the DNA is reliant on the DNase I step performed after RNA product is complete. The EMA document suggests sizable (10×) batch-to-batch variation in the amount of residual DNA present (10-220 ng/mg) in production batches, even if all of these were below their specification of 330 ng DNA/mg RNA. One development batch did have much higher template concentration (815 ng/mg) due to an "incorrect DNase I stock solution", as you noted.
3) The assay to detect residual DNA is a qPCR-based assay with primers on the T7 promoter and Kozak sequence. Presumably the motivation for this assay is high sensitivity to detect what is expected to be a rather trace impurity. This method will not quantify any DNA not containing the target sequence, but should detect the input plasmids sequenced here (I checked and the primers do match).
4) The residual template check is done after the Drug Substance production stage, prior to the LNP Fabrication stage.
5) Key point: After this stage, no further testing for residual DNA is performed. The amount of residual DNA is assumed to be within spec after that in the initial test. Many of the methods used to check RNA will happily detect DNA as well, such as the RT-PCR you highlighted. I have found no information on the amount of DNA present in the batch QCs reported in the TGA FOI requests (e.g. FOI 3471 from https://www.tga.gov.au/resources/publication/publications/documents-released-under-section-11c-freedom-information-act-1982-jul-2021-jun-2022).
6) The is an oversight in this process that I can see, which is there is no guarantee that the subsequent steps used in product will not effectively concentrate the DNA relative to the RNA. For example, the DNA will be obviously a lot more stable than RNA, so will be more resistant to degradation. In addition, what if the LNP is more efficient at packaging DNA than RNA?
The last point could alone potentially bring the residual amount of DNA to above the specification of 330 ng/mg RNA, if it was already close to the spec. I was trying to dig up some figures on what the RNA losses are going from Drug Substance production to final product to estimate worst case scenario but haven't found any yet. Nevertheless, I assume the losses are <25% at worse case, so in order for a large amount of DNA to be present the DNAse would have to fail, and the residual DNA assay would have to fail or be ignored, implying extremely shoddy standards. In any case, residual DNA testing should really by part of the QC for the final product, but it's not. That's lax in itself.
As for the specification itself, the EMA document has this justification:
"The specification for residual DNA template was based on the WHO recommendation of not more than 10 ng DNA/dose. Based on these considerations, and assuming a maximum dose of 30μg, the commercial acceptance criterion at release is ≤330 ng DNA/mg RNA. The approach is endorsed. "
I haven't looked at the WHO justification for this and I'm not sure I really agree 10 ng is acceptable. This is more than enough DNA to transfect some cells, with potential genomic integration, when multiplied over enough cells exposed and enough recipients.