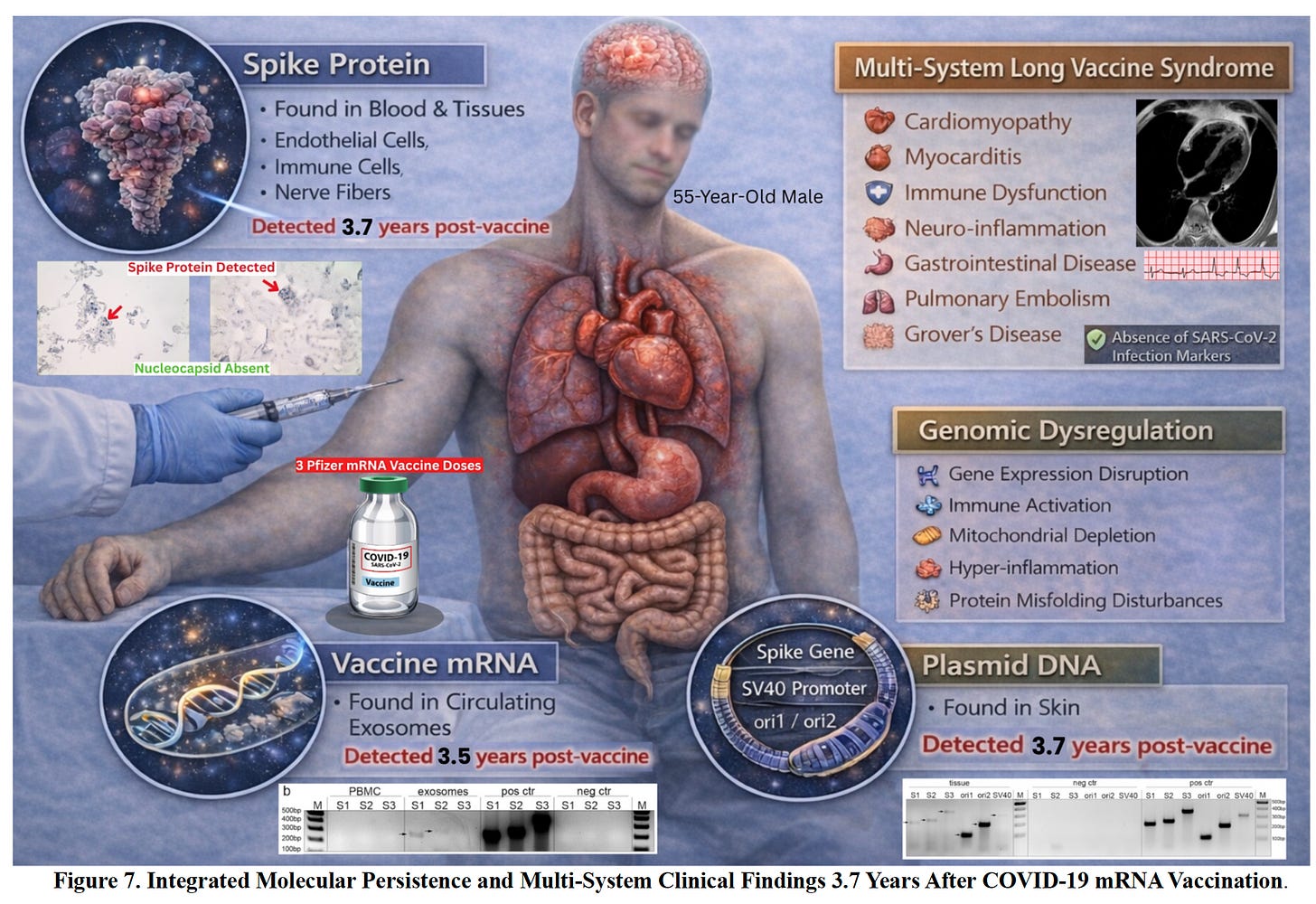
Spike Protein, RNA and DNA record breaker
Nicholas Hulscher et al
I was asked my thoughts about this preprint paper.
It should be known that I am biased. I’ve worked with these authors before and have deep respect for the risk they are taking doing this work and how hard it is to get it done.
I’m going to limit my comments to my speciality of DNA and RNA detection as I think we can learn a few things as a community on how to interpret data like this. I have not scrutinized the complete paper yet. I focused in on the questions people asked of me.
First thing everyone has to appreciate when looking at PCR of Spike in a human background is that Pfizer and Moderna codon optimized their Spike sequence and did so very recklessly.
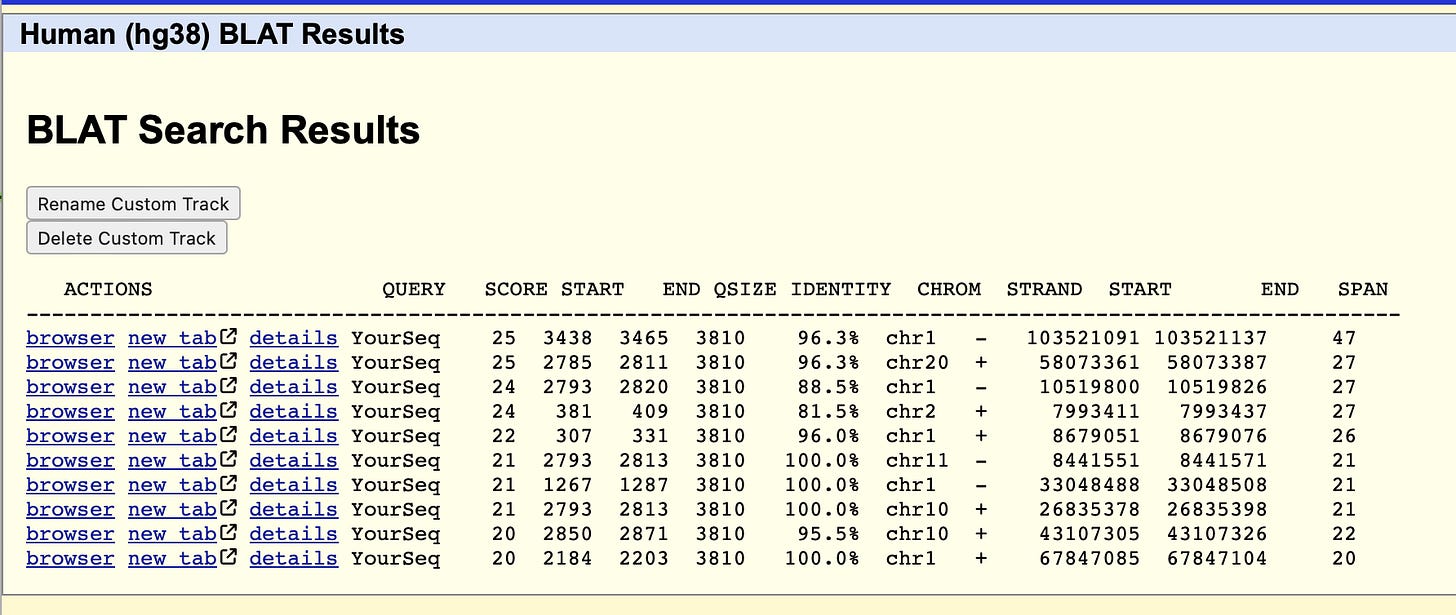
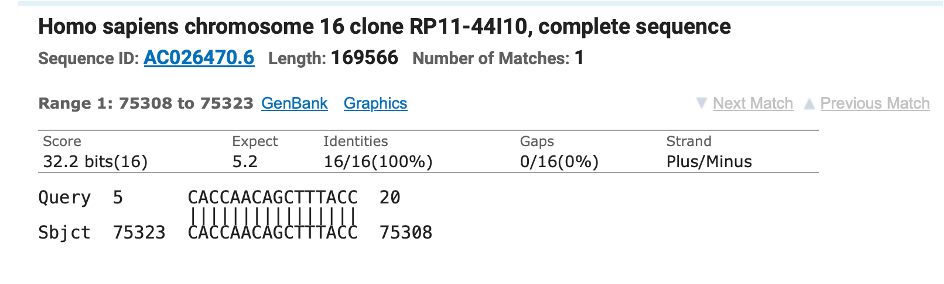
If you take the Pfizer Spike sequence and BLAT it against the human genome you can find many regions in the vaccine that map 100% to the Human genome for short 20-47bp spans. This means primers that perfectly amplify vaccine vials, may cross react with human DNA and cause false positive PCR signals when human DNA is in the background. They may be clean in vaccine positive controls but produce unexpected bands once applied to human DNA looking for vaccine DNA.
This alone should scare the shit out you as there is something known as RNA interference that will process mRNA like this and down regulate the genes that have homology to this spike sequence. It requires about 20-22bp to trigger.
I have a whole thread on this mess here:
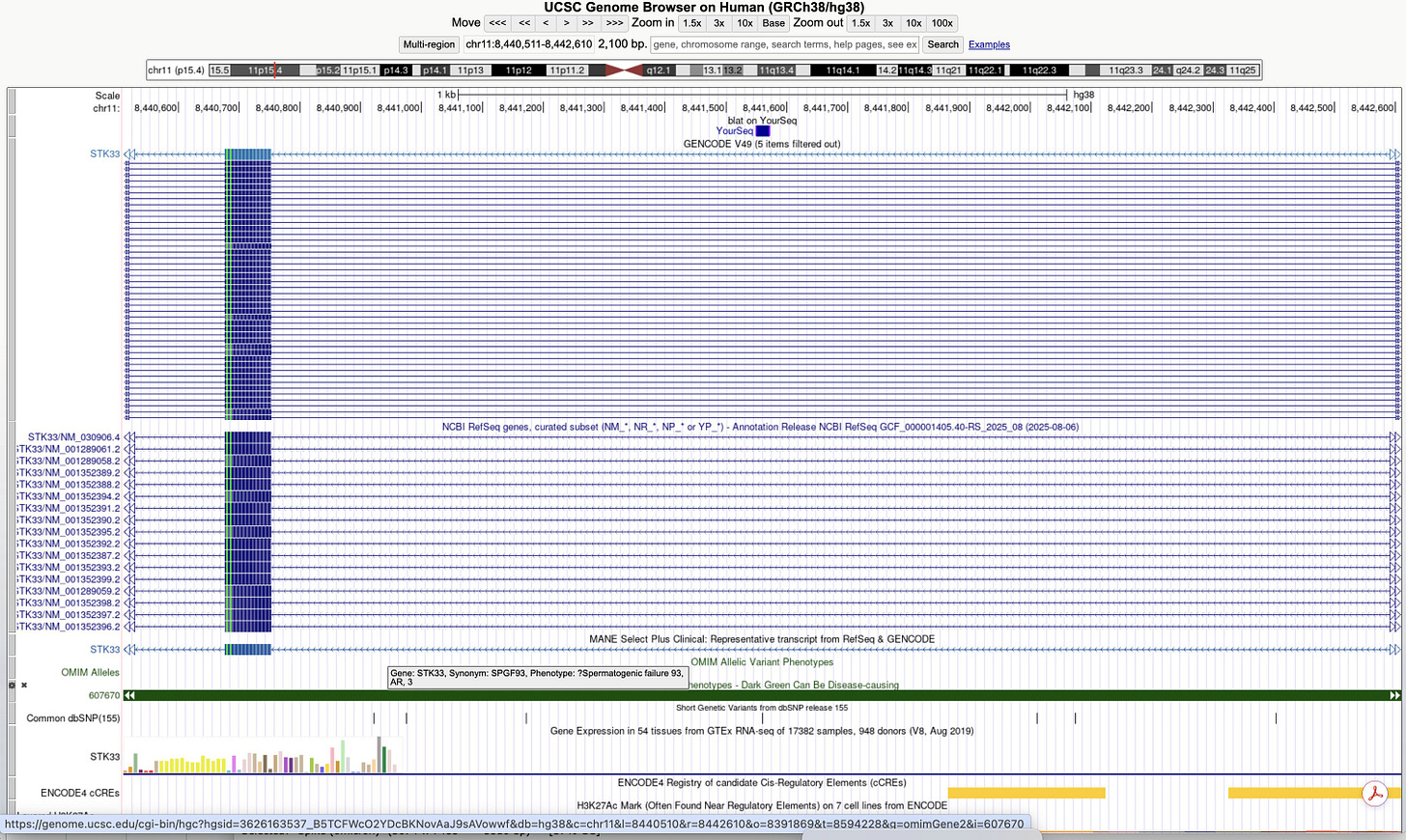
Some of these sequences are perfect hits to OMIM genes involved in Spermatogenic failure and we have evidence of that actually happening in the vaccinated.
OMIM genes are the genes best nailed down to be causative of disease.
To make matters worse, several of the hits in the BLAT table above are to genes expressed in testis.
The viral spike doesn’t have this homology. Its purely a product of Pfizer “codon optimization”.
Many of these studies looking at sperm motility/count obscure this signal by mixing Moderna and Pfizer patients but if you only look at Pfizer, which uniquely has this homology, you can find the problem in Gat et al.
So you need to take the primers used in any paper like this and run them through NCBI-primer BLAST against the Human genome and human transcripts.
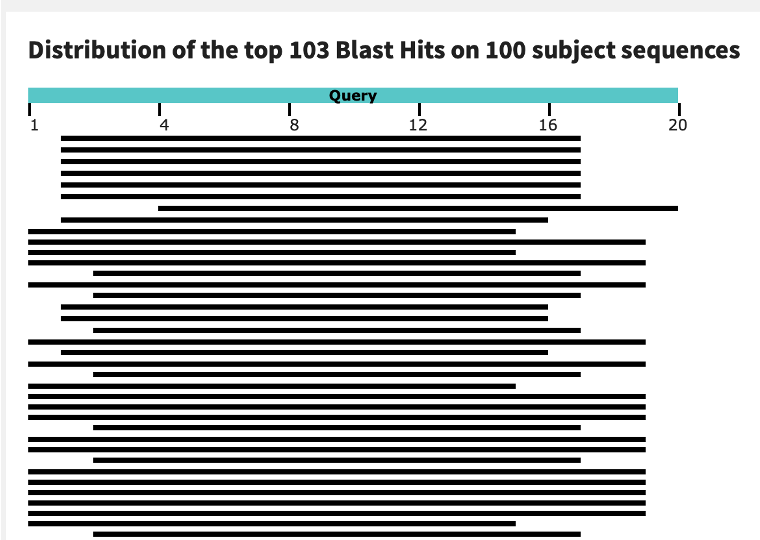
It’s important to use the Primer-BLAST tool. If you just BLAST primers as single sequences, you will get lots of short hits to such a large genome but the only hits that matter are when both primers are in close proximity in the genome as PCR doesnt work across chromosomes and is usually limited to under 4kb. With perfect conditions, you get ~20kb to amplify with very specific enzymes and primer design.
Many people took the simple BLAST shortcut to attack the last paper that found this as these people have zero experience making PCR assays.
The NCBI-Primer BLAST takes into consideration the orientation and distance of each primer to give you a better sense if the amplicons are possible.
For example, here is one of Hulschers Spike assays. It can make a 102 bp amplicon on Human DNA if you allow for 3 mismatches in each primer. The 1224bp amplicon, Im less concerned about as their bands on the gel dont imply this is happening.
One limitation of the study is that they are not using qPCR which requires 3 primers to all be properly arranged and it gives you a quantitative framework. Primers that amplify at CT 35 in human are less convincing than primers that fire off at a CT of 25 (1000 fold more signal).
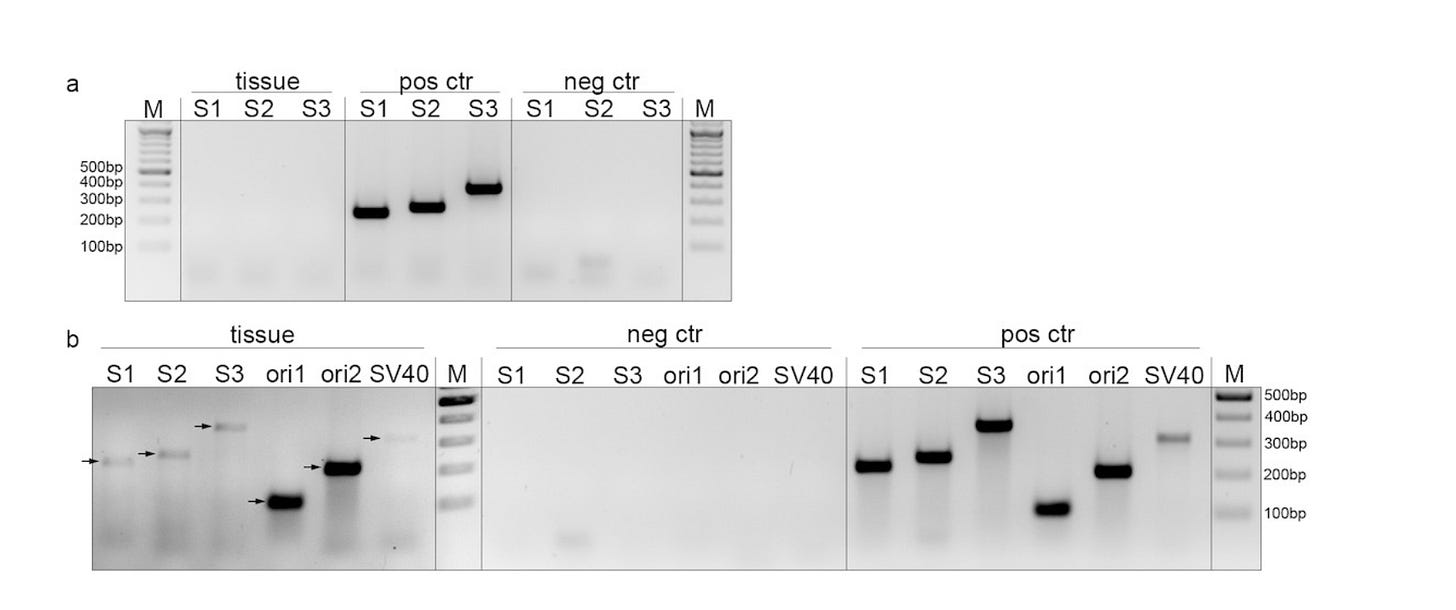
So when you have just bands on a gel, where the cycling conditions are not specified, you have to sanger sequence them to confirm. They did this for the Ori primers and they match Pfizer but there doesnt appear to be spike or SV40 Sanger confirmation.
Ori is its own worst enemy. This sequence contaminates nearly all PCR polymerase kits as the polymerases are usually expressed in E.coli with a plasmid that contains this Ori so its a well known Kitome contaminant. These kitome contaminants usually only light up after CT 30 but we dont have this information from the Hulscher paper.
They rightly don’t make bold claims about Ori amplification and acknowledge it could have different sources.
I think the most compelling figure in the paper is 6B.
When you see 6 amplicons all popping up in the tissue but not that negative control, it becomes harder to believe these are all off target as I cant find off target hits for S1 or S3 primers using NCBI-Primer BLAST tool.
The Ori, SV40 and S2 primers I can find some human primer hits, albeit with mismatches and which need further scrutiny as NCBIs human genome is often contaminated with these vector sequences which are from the sequencing vectors people use in sequencing projects. They are mislabelled as part of human cDNAs because those cDNAs were sequenced from plasmids which contained those sequences and they were not fully removed from the database. I have not done this level of forensics as Figure 6B looks to have all 6 amplifying, albeit at very different intensities but of the expected size seen in the positive controls. Given all of the other clinical data they have corroborating spike expression, this is likely real.
It would help if they better described their negative controls? Are they water or are they human DNA pre-pandemic? The methods section on the PCR conditions needs more detail. How many cycles etc…
You will likely see the typical critiques of the paper where some newb just BLASTs a single primer and gets human hits like this and declares “FRAUD”. US Mortality beclowned himself once like this before.
Ignore these attention seeking amateurs. They don’t understand the primers need to match on the 3’ ends and be pointed at each other <4kb apart for any amplification to actually occur. The paper they critiqued claimed to have sanger confirmed the amplicons such that these critiques were shallow gotcha’s. That paper was mysteriously taken down and was by no means perfect but the primer concerns raised were shallow single BLASTs like seen below. One 15bp homology or even two on different chromosomes are irrelevant. Not ideal but not fatal.
If the critique comes from the PubSmear mob..
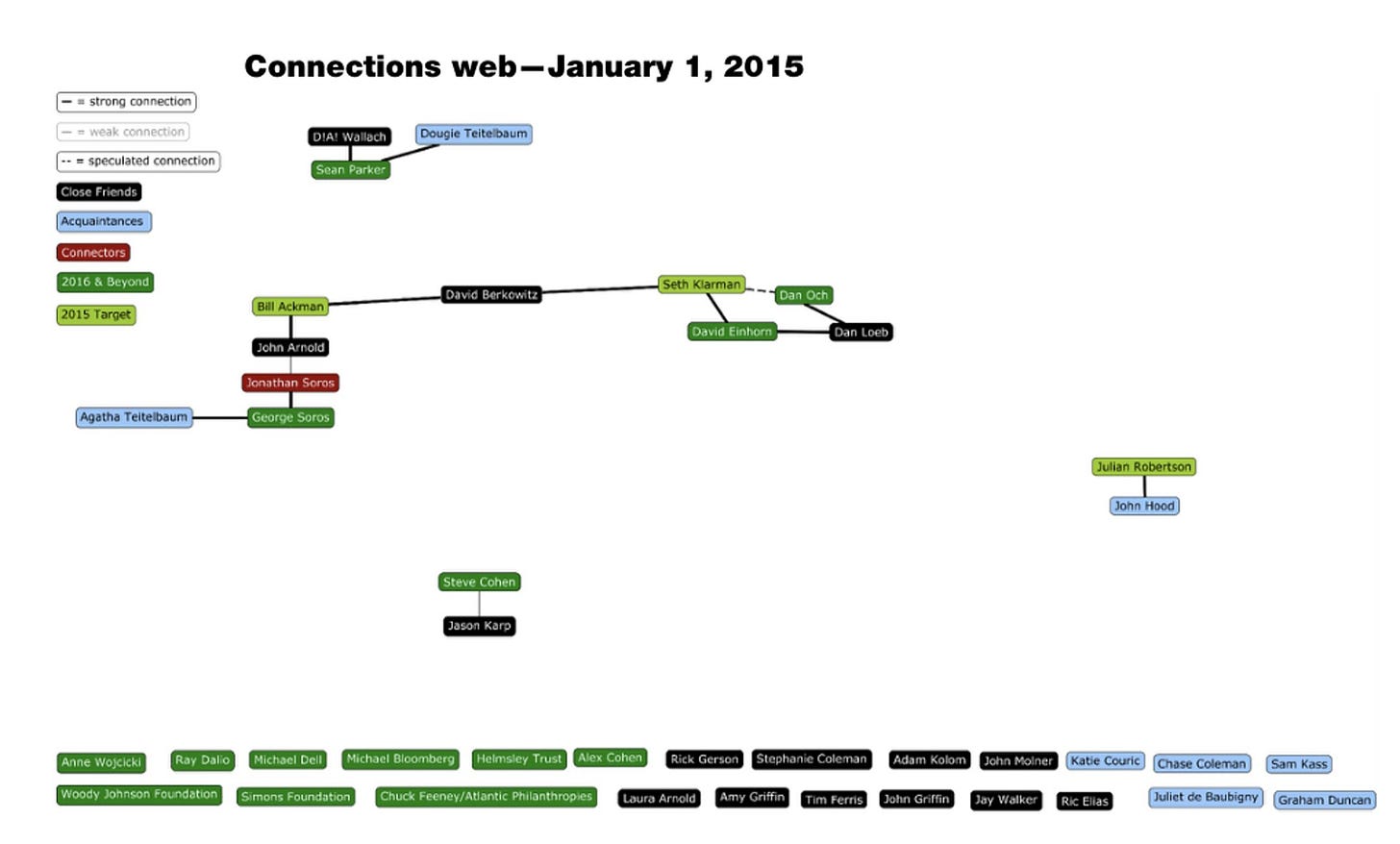
Just remind them that their funders (Arnold Foundation) are Close Friends with Jeffrey Epstein and they should pack their bags. No one should trust them anymore.
You read that correctly. The Epstein docs are live and there is a contact map published that has John and Laura Arnold as “Close Friends” of the Epstein mob. And its is no surprise we can see emails inside that network planning for Pandemics to capitalize on them. I’ve seen many super hero vs villains films and none of the movies had villains this evil or bereft of ethics. Truth is stranger than fiction.
This is a case where you have to take on the totality of evidence. They have Spike protein detected years later and some evidence of mRNA and plasmid DNA in particular cells or tissues of interest. Also gene expression changes and multiple labs confirming this. Anyone datapoint you might be able to contest but all them together make a pretty compelling story. This is a preprint. Its likely to clarify some of these details in final publication but important to get out ASAP.
It would be very interesting to see if their RNA-Seq data supports the pathways we expect to be dysregulated by these contaminants (cGAS-STING, IL-10, RIG-1, TLR7/8/9) recently highlighted in Dr. Robert W. Malone substack on this.
If I were reviewing this, I’d suggest
1)Better describe the NTCs? Are these water or pre-pandemic human negatives. How do these primers behave on Corielle human pre-pandemic DNA controls like NA12878?
2)Better disclosure on PCR cycles, conditions etc.
3)Might want to use DNaseI-XT for your RT-PCR as DNaseI doesnt process the spike DNA very well (McKernan et al). Not critical for publication but maybe for future work.
4)Any Sanger data on Spike S1, S2, S3? Bands are dim but perhaps nested PCR can rescue these?
5)Perhaps supplement disclosure of the NCBI Primer-BLAST hits for the primers. I’ve run all 6 through and they don’t look as bad as what might occur when someone just single primer BLASTs these primers.
6)Don’t be afraid to self cite. The Kammerer et al paper is critical context for this. The SV40 cell passaging work adds to the credibility of this observation. PubSmear loves to critique on this axis but they are just wrong in this case.
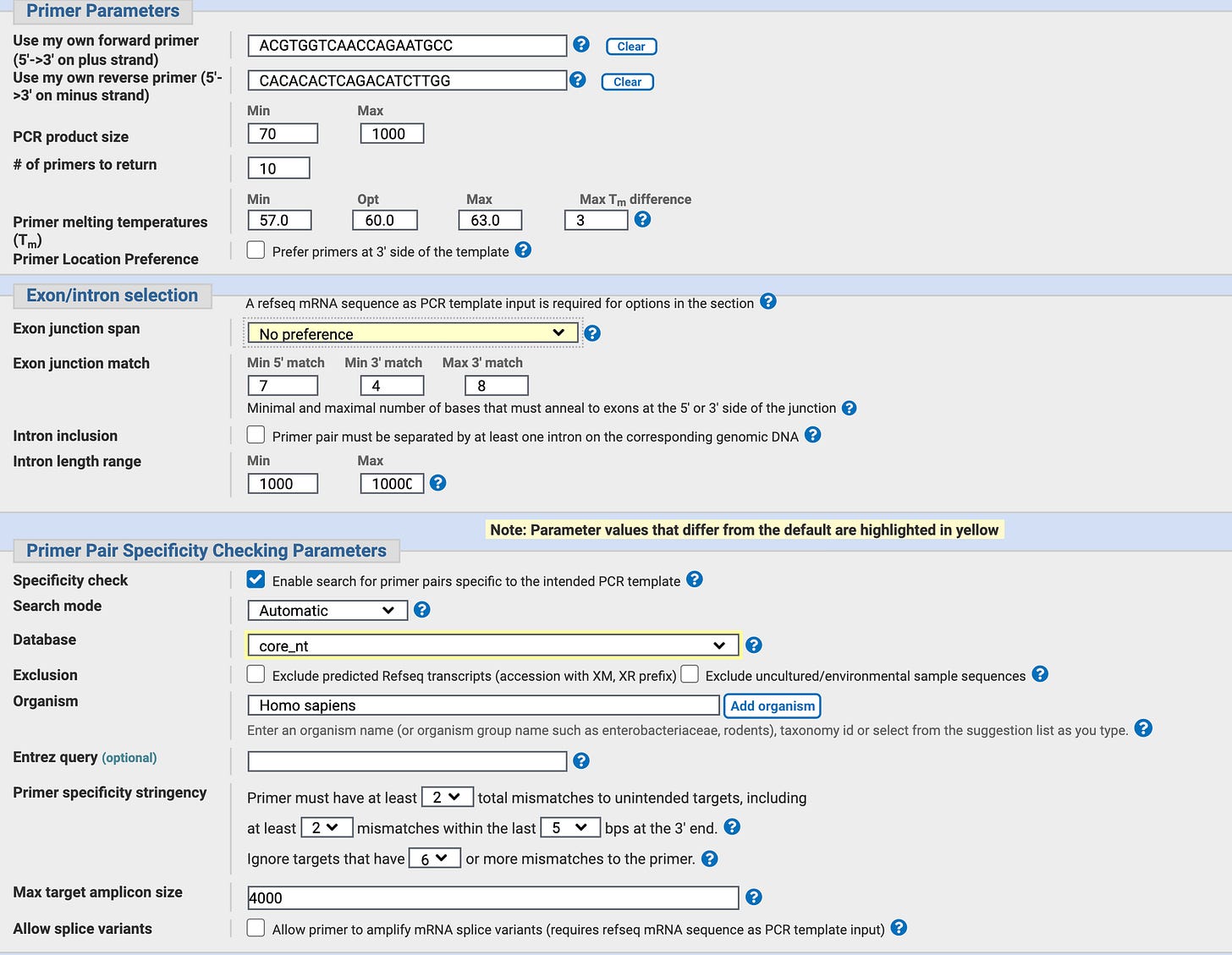
Some of the parameters used in NCBI Primer-BLAST