Are longer dsDNA fragments present in BNT162b2?
Dr. Sin Lee further explores the dsDNA contamination
Dr. Sin Hang Lee, MD, F.R.C.P.(C), FCAP of Milford Molecular Diagnostics designed his own BNT162b2 amplicons targeting longer molecules for PCR and Sanger sequencing. This is an important evaluation to ensure there is nothing magical about the primers we designed for qPCR. Dr. Sin Lee has already sequenced amplicons generated from the BNT162b2 qPCR assays we designed and published earlier.
Independent primer design, amplification and sequencing from the Illumina consensus sequence we produced is more independent validation of the initial results.
Dr. Lee used a 2 step process for amplification using a unique high fidelity polymerase. 30 cycles of PCR amplicons were run on a gel and then re-amplified with another 30 cycles using the same primers.
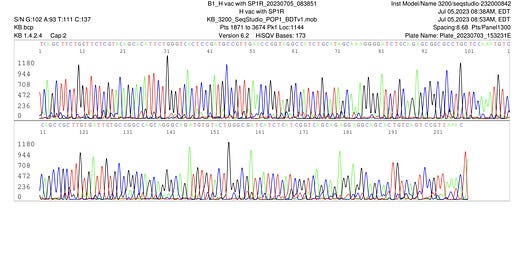
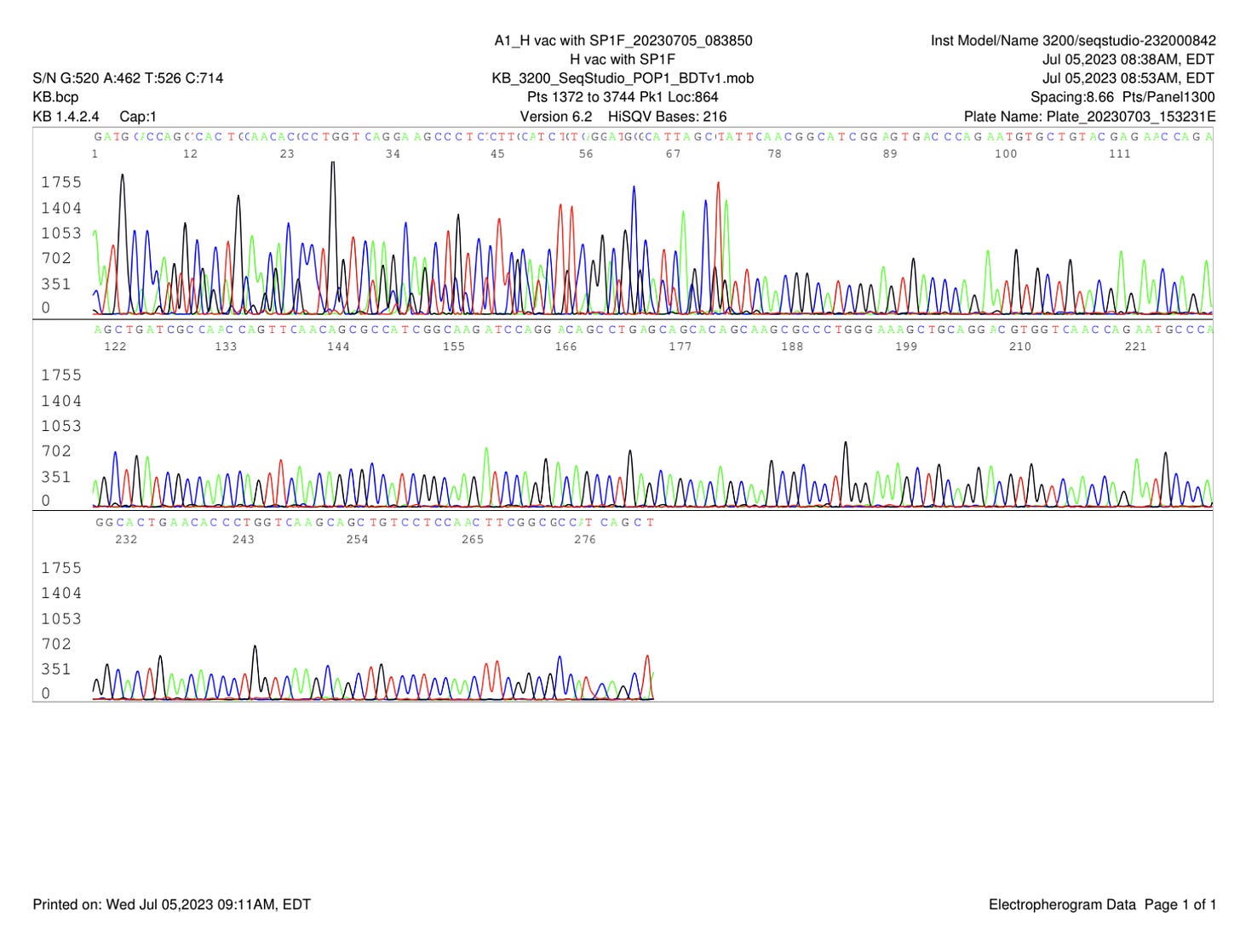
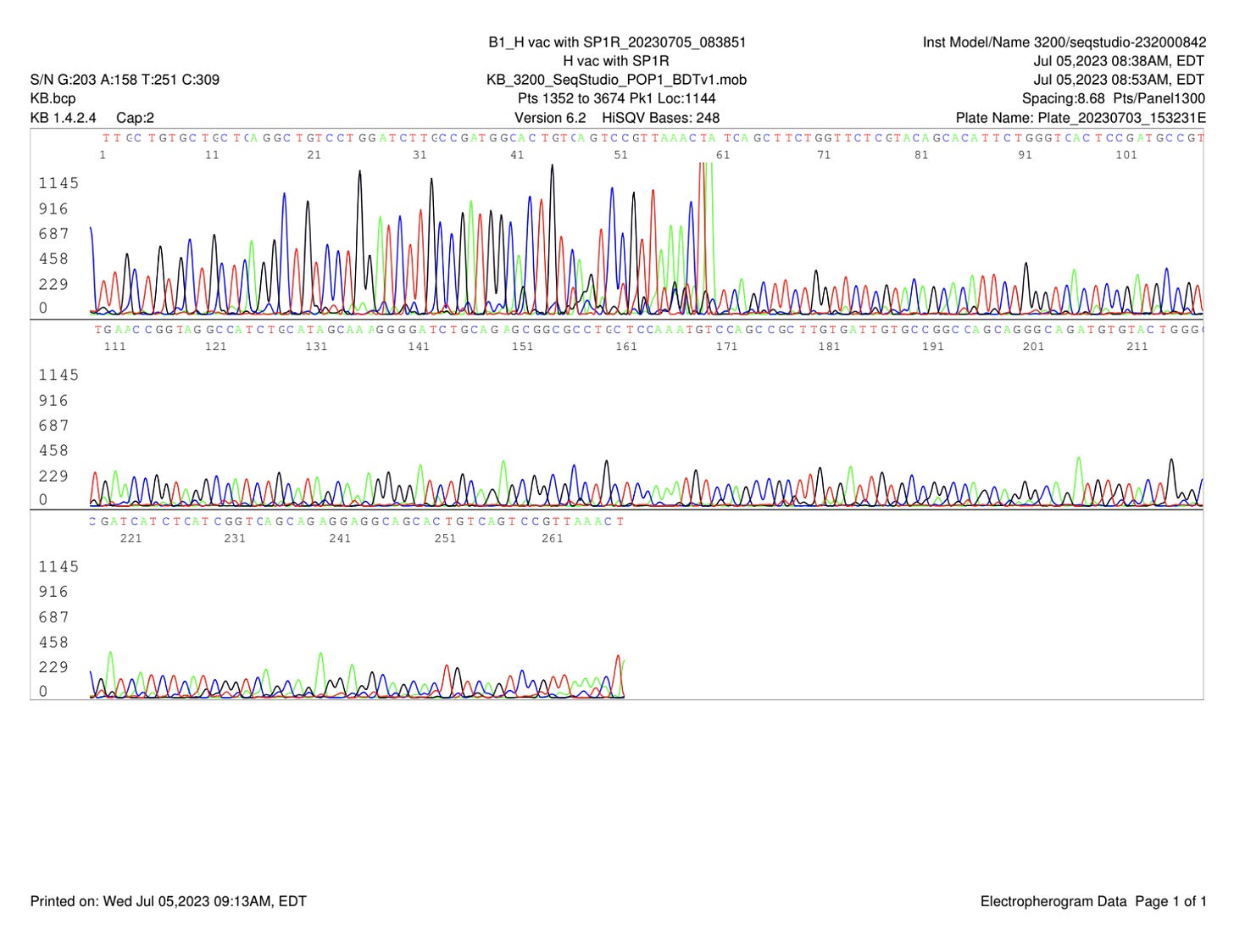
Some smaller off target amplicons are seen after 60 cycles (Figure 1). These can be seen obscuring the Sanger sequences 1st ~70bases of sequence (Figure 2). Dr. Lee did not perform a DNA purifications step to eliminate the smaller amplicons.
Trimming of the high quality sequence is shown in Figure 3.
This cleaner/trimmed Sanger sequence represents this region highlighted in Blue/Green of the Spike sequence from Figure 4.
The Primers used for amplification are seen in figure 7.
The quality trimmed Sanger reads were assembled and BLAST’d against NCBI to find a 100% alignment to a 363 bp region BNT162b2 spike sequence (Figure 8).

Dr. Sin Lee’s comments on the small off target amplicons shown in Figure 9.
Dr. Sin Lee further explored if his primers had any off target locations in the vector sequence. Off target locations like this could result in these smaller amplicons when using high cycles of PCR. With 60 cycles of PCR where the primers are not nested (same primers used for both series of 30 cycles in Non-Nested PCR), one can often find these off target amplicons that obscure some of the Sanger sequencing. However the assembly of these sequences results in a consensus that matches 100% to 363 bases of the spike sequence.
It’s important to note that no DNA purification was performed to eliminate small amplicons before sequencing. While the small amplicon is dimly seen on the agarose gel in Figure 1, intercalating dyes stain DNA with increasing intensity based on their size. Smaller amplicons stain less intensely. Electrophoretic injections involved in Capillary sequencing also favor smaller amplicons. This explains the higher intensity signal observed on the smaller amplicon (Figure 10, 11, 12). Despite, this quality trimming with forward and reverse reads provides enough high quality sequence to span the 363bp amplicon.
This is an important form of validation as some of the guidelines speak to fragments over 200bp. Our qPCR amplicons are 105 and 114bp and may over quantitate the dsDNA 200bp and larger. Seeing 363 base fragments amplify and sequence demonstrates there are DNA fragments over 200bp in the BNT162b2 vaccine but the quantitation of this size of the library still requires further measurement.
For reference, Dr. Lee’s primers flank the amplicon we designed and provide independent verification of the primer sequences used in previous studies.












Now *THIS* is REAL science. Dr. McKernan presents us with a thorough description of Dr. Sin Lee's methods and materials, makes the raw data publicly available, with minimal discussion. Great work that deserves to be saved, shared, and analyzed. Thank you, Dr.s, for your hard work and genuine contributions to Humanity.
Thank you Kevin McKernan for your work that has provided the wold with great advancements in understanding the molecular biology of the construct, and for keeping us updated on the related contributions of others.