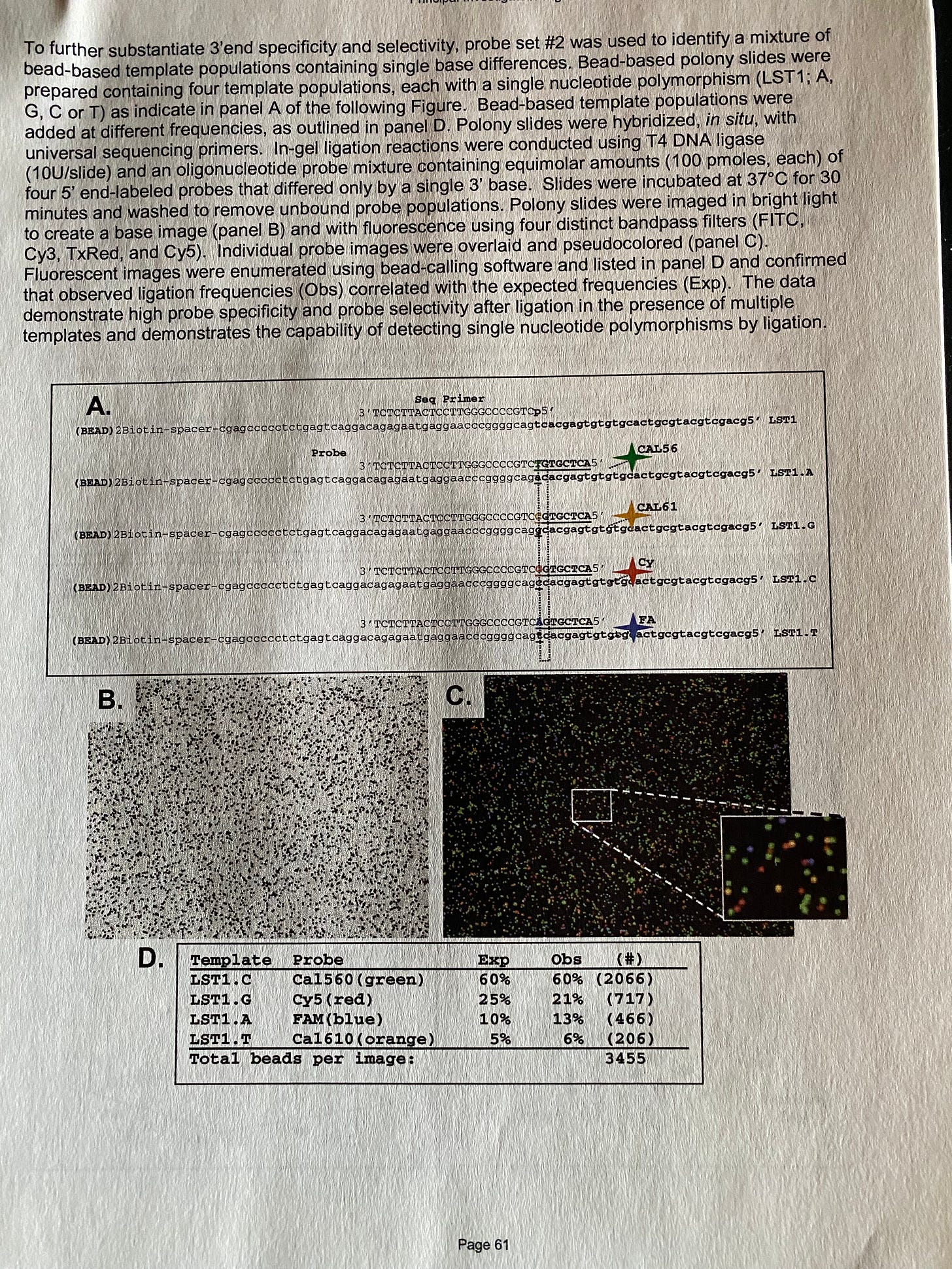
Jessica Rose posted a great review on DNA sequencing.
I went over this topic on a video presentation with PANDA ~10 months ago.
There is a great book on this race to $1000 genome by Kevin Davies.
Jessica is spot on in her description. This revolution was enabled by permissionless innovation and competition. NHGRI likes to take a lot of credit for this advance. They did in fact fund a lot of labs to develop these tools (including our lab) but the irony in that funding is that the companies with the least amount of NHGRI funding took the majority of the market share (Illumina). This is perplexing and not obvious to those on the sidelines.
I’ve written about this before. People like to point to NIH funding as an example of the government addressing ‘Market failure’ and investing stolen dollars to where a select group of people believe its best allocated. This select group of people often called a ‘NIH Study Section’ is generally made up of very accomplished people from industry and academic centers but they are just a subsampling of the 10-12 influential minds in the field. They do not encompass the entire intellect of the field and thus are a subsampling suffering from the economic calculation problem described by Ludwig von Mises.
The argument often goes like this…
Since the Private sector is greed based, they never invest in high risk/high return projects due to quarterly earnings pressure. Therefor the government needs to steal money from everyone (including the people in the study section) to invest in Space programs that create derivatives like Tang.
This is false. There is no way to capture an NIH grant without preliminary data. In our case with the SOLiD sequencer, we had to have $3 Million dollars worth of preliminary data in order to be awarded a $6M grant.
To give you some perspective, I’ve included parts of the grant application. You can see we had 90% of the proof of concept worked out before we submitted the application. The rest was engineering. This is why im on the same page with
regarding the DEFUSE proposal. You dont submit such grant applications with no preliminary data and expect it to get funded. The application require 100s of pages of work and no one wastes their time on that process unless they have nearly 90% of it already worked out and thus present a very low risk option for the NIH to parade in front of.As many innovative projects will tell you, we shifted gears often and this was difficult to do if each change required permission from NIH. We shifted gears from reversible terminators to Ligation based sequencing as it was making more progress but this change in direction required we update our grant application to get approval for the change in direction. We could not afford to do this for every change we thought of so some of the changes were funded by private dollars while others were spent with NIH grant funds.
We also ditched sticking beads to glass with acrylamide gels and went with a direct coupling to glass. We tweaked everything we had in the initial application as we discovered what worked and what didnt work.
Prototypes were fun to build.
In some circles of the internet I have been accused of being a chemist. Im not trained as one but I was able to read very interesting work from Kool et al and realize it enabled a new form of DNA sequencing. By replacing an oxygen on the 3’ end of DNA with a Sulfur group, we made a new class of reversible terminator oligos. This Phosphothiolate linkage is cleavable with aqueous silver. We could ligate in 8 base probes with dyes and cut the dye off after imaging at base 5. Thereby sequencing DNA in 5 base hops.
We of course had to prove this with model templates run on traditional Sanger CE instruments.
I wish I had frequent flier miles at IDT as we ordered hundreds of thousands of dollars of labelled oligos from them over the course of this project testing out various perturbations we could do to the oligos and how the ligases would handle them.
We had to clone many of the products using these ligation methods and sequence them with Sanger sequencing to understand the fidelity of T4 DNA ligase.
Next, we had to explore different fluorescent dyes to see if the ligases had any preferences.
Getting the chemistry to work was half the battle. We had to image with submicron resolution 100,000s of image tiles per run.
We had to do this in 4 channels and spectrally separate the overlapping spectra of the dyes. This had to be done quickly. Each sequencer had 20Tb of Disk and a $25K compute cluster just to keep up it. This was before GPUs had made this much simpler.
All this had to be affordable and show a roadmap below a $50K genome. We eventually got it below $2K with ABIs helps.
So who funded all of this preliminary work required to be awarded an NHGRI grant?
The greedy private sector that is ‘incapable of High risk, High return’ investment! Market Failure! Sure, the NIH grant helped to reassure our investors that we were not nuts and the investors loved the idea of non-dilutive government money fueling our R&D pipeline.
But the riskiest bets were made 2 years before the NHGRI chipped anything in. We had to hire people, buy equipment, build prototypes, license patents, synthesize reversible terminators, test them, scrap them, redesign for ligation based sequencing and build software and computer systems that could keep up with this torrent of data.
We had about 30 ABI 3700s and 3730xls running at Agencourt at the time. Each was a $350K instrument that required a $1-2M robotics pipeline to feed. Every SOLiD instrument we built was the equivalent of 5,000 ABIs. This was more capacity than the world had with a single instrument.
This really struct home when we finally had 10 SOLiD sequencers humming along in Beverly Mass performing 50Gb runs and in a week we generated more sequencing data than was in all of Genbank from the beginning of time!
The marketing folks couldn’t predict the market size as the world had never seen such sequencing capacity before and generally under estimated the revolution ahead of us. The small minded marketing executive would tell us to make the instruments smaller and slower so you don’t cannibalize the sequencing market with a single machine.
The opposite was true. The lower we brought the cost of sequencing, the more people discovered and the more hungry they became for more sequencing. There continues to be an insatiable appetite today.
So why did Illumina dominate the market? Several reasons. ABI was in the stage of Sanger Sequencing where they enjoyed a decade long monopoly on Sequencing and were thus in the harvesting era of their technology. They had outsourced all of the manufacturing to Hitachi as the cost of engineering in Silicon valley was a drag on the P&L. This is not uncommon late in a technology life cycle. Its called harvesting as they are cutting costs, increasing margins and preparing for the next wave. This created a large engineering flight out of the company to Illumina and PacBio.
When we realized we were outgunned by the engineering team at Illumina, we tried to repeat this ‘harvesting plan’ by outsourcing the engineering of the SOLiD 5 to Hitachi. All of this complex chemistry had to be translated into Japanese only to watch the 2011 Tsunami take out the SOLiD factory in Japan.
This was the final blow for SOLiD but it only captured ~30% of the market share before that. One of its limitations was that we engineered Mate paired DNA sequencing instead of paired end sequencing. The former required 10 fold more DNA but offered very unique long range (40Kb) paired end reads. We eventually got paired end sequencing to work but the reverse reads were limited to 35bp reads.
While we had log scale better accuracy than Illumina, our read length limitations and late arrival of paired end reads made the platform less appealing.
Mate-pairs were something we had grant money for and thus it was easy to tell ABI that we should finish the grant goals on Mate-pairs and tackle paired ends later. This was an example of obsolete Study section guidance. The market was begging for paired ends but we were funded to investigate mate pairs as that seemed like a good idea 3 years ago. ABI eventually cancelled the grant as it was too small and required IP disclosures and different GAAP accounting. This was a smart move as it focused the group on their needs and not an outside study section.
SOLiD also had a unique way of reading DNA (2base encoding) that required we write the majority of bioinformatics code for handling SOLiD data. Illumina had the benefit of the community writing the code for them as their single base encoding was more conventional. The SOLiD platform really needed the 2 base encoding to clean up the ligation error but there were also IP landmines pushing us in this direction.
So in summary, Grant funding often locks your development team into satisfying grant objectives that are frequently detached from the immediate market needs. The voice of the customer should drive your R&D not a study section of smart people with no skin in the game. Other large NHGRI grantees suffered similar fates from 454 to Helicos.
The argument for Market Failure and graft based funding is clearly false. No one can apply and succeed in being awarded an NIH grant without preliminary funding from another source. Like Elizabeth Warren, they will claim, “You didn’t build that. We did” and they will be lying once again as many unaccountably government agencies often do.
Yes, I much prefer government graft being spent on DNA sequencing than bombing the Middle East. But this is a very slippery slope once you realize similar funding bodies were involved in funding GOF at WIV, fabricating Proximal Origins papers, censoring
, Martin Kuldorff, Sunetra Gupta, while having overt conflicts of interest in mRNA vaccine royalties while they demonized generic drugs… that this market failure argument is far from Safe and Effective.
































What a great article exposing one of the many hypocrisies of the NIH.
Well done for taking the time and effort of writing this, Jessica too for the intro.
"This is why im on the same page with Charles Rixey, MA, MBA (c) regarding the DEFUSE proposal. You dont submit such grant applications with no preliminary data and expect it to get funded. The application require 100s of pages of work and no one wastes their time on that process unless they have nearly 90% of it already worked out and thus present a very low risk option for the NIH to parade in front of."
I never heard anyone make that point, but now you explain it, it seems obvious. Very important.