There have been many concerned researchers who have posted about the poor RNA integrity scores (RIN) that occurred in the switch from Process 1 to Process 2 manufacturing with Pfizer. Many have been worried about what these ‘wrong size’ modRNAs represent.
and have both covered this. We also covered this concern in our PrePrint with .The N1-methyl-pseudouridine is a base that radically alters the Tm of the nucleic acid and as a result the T7 Polymerase that is making copies of this modRNA gets jammed up like a vacuum trying to recoil its twisted power cable. In this process of T7 polymerase copying the DNA into modRNA it can flip strands and make chimeric modRNA.
This is also known as template switching in the literature.
Jikkyleaks covers the dirty RIN plots on X.
The EMA documents made note of a massive decay in RIN scores (RNA Integrity) when switching from Process 1 to Process 2 manufacturing with Pfizer.
So what is Strand flipping, also known as template strand switching?
A schematic that helps visualize cis-template switching in A, and trans-template switching in B.
Another way to view this below
What does this look like on a radio-labelled nucleotide, T7 polymerase extension?
Moderna has that answer for you in their paper from 2023 when they engineered a mutant T7 polymerase that does less of this (G47A +884G).
Note the reduction of dsRNA in the vaccine as a result of this below (Red = mutant, black = WT T7 polymerase). This is what is causing the low RIN scores. It’s not RNA decay, it’s transcriptional fidelity issues.
So how do we measure the dsRNA in the modRNA vaccines made before this new T7 polymerase was invented?
RNA sequencing on Illumina can be performed in a such a way that the strandedness of the mRNA is retained in the DNA sequencing data. What does that mean?
Most expressed mRNA is Sense strand and single stranded RNA. This is also known as the Crick strand. This is the strand that is coding for a messenger RNA. This gets confusing in the Pfizer case as both strands code for a message in the spike region and only one should. Read the previous stack and you will learn that the Pfizer plasmid has an undisclosed spidroin Open Reading Frame on the opposite strand of Spike. We’ll refer to the intended Spike coding strand as the Crick strand.
The antisense strand is known as the Watson strand. Only DNA has both Watson and Crick strands. mRNA should only have Crick (sense) strands as its single stranded coding message.
Most RNA sequencing generated today is in fact generated on DNA sequencers. Oxford Nanopore is an exception but their Direct RNA sequencing is still nascent. We would use ONT but the N1-methyl-PseudoU screws up their base caller and needs more work.
So in order to use most other DNA sequencers, all RNA must first be converted into DNA in order for it to be sequenced. Once you convert this into DNA you have 2 strands and we want to ensure we don’t confuse ourselves over which one of those strands was the original strand in the sample. This is important if we want to quantitate how much template switching is going on in the vaccine. We have to carefully itemize the origins of each strand.
The method we used to capture stranded RNA libraries is from NEB. They have a clever method that 1st converts the single stranded RNA into single stranded DNA. This first strand synthesis step is supposed to favor RNA over DNA with the use of actinomycin D. This additive is explained in a previous stack. Its an intercalating inhibitor of RNA polymerases that disfavors this enzymes amplification of double stranded molecules and favors the amplification of ssRNA and ssDNA.
The second strand of DNA is then synthesized with a removable base (DNA based Uracil). This DNA will have A overhangs on the 3’ end of each strand. Once you have this plusA dsDNA, you can use a double stranded ligase to glue (ligate) hairpin primers with Ts on specific ends to direct the the dsDNA ligation in a very strand directed manner.
The bottom Watson strand can then be removed with UNG which digests DNA with Uracils. Now the only strand that amplifies has your sequencing primers on both sides with the Green Primer on the 5’ most end of the Crick strand and the yellow primer on the 3’ end of the crick strand.
You can then sequence these DNA strands from both Ends (paired end sequencing) and count reads that are from Crick vs Watson strands and begin to quantitate the template switching frequency.
There are tools online that can help you sort these sequencing reads into the proper buckets of Watson and Crick sourced templates.
The shell script below requires samtools to be installed. In the past I had shared some strandedness plots but they were not analyzed on the finished NCBI references and have artifacts. Ignore those. These are analyzed using scripts in common use in the field and mapping the reads to the right references.
Using this shell script for teasing apart the strandedness of the reads.
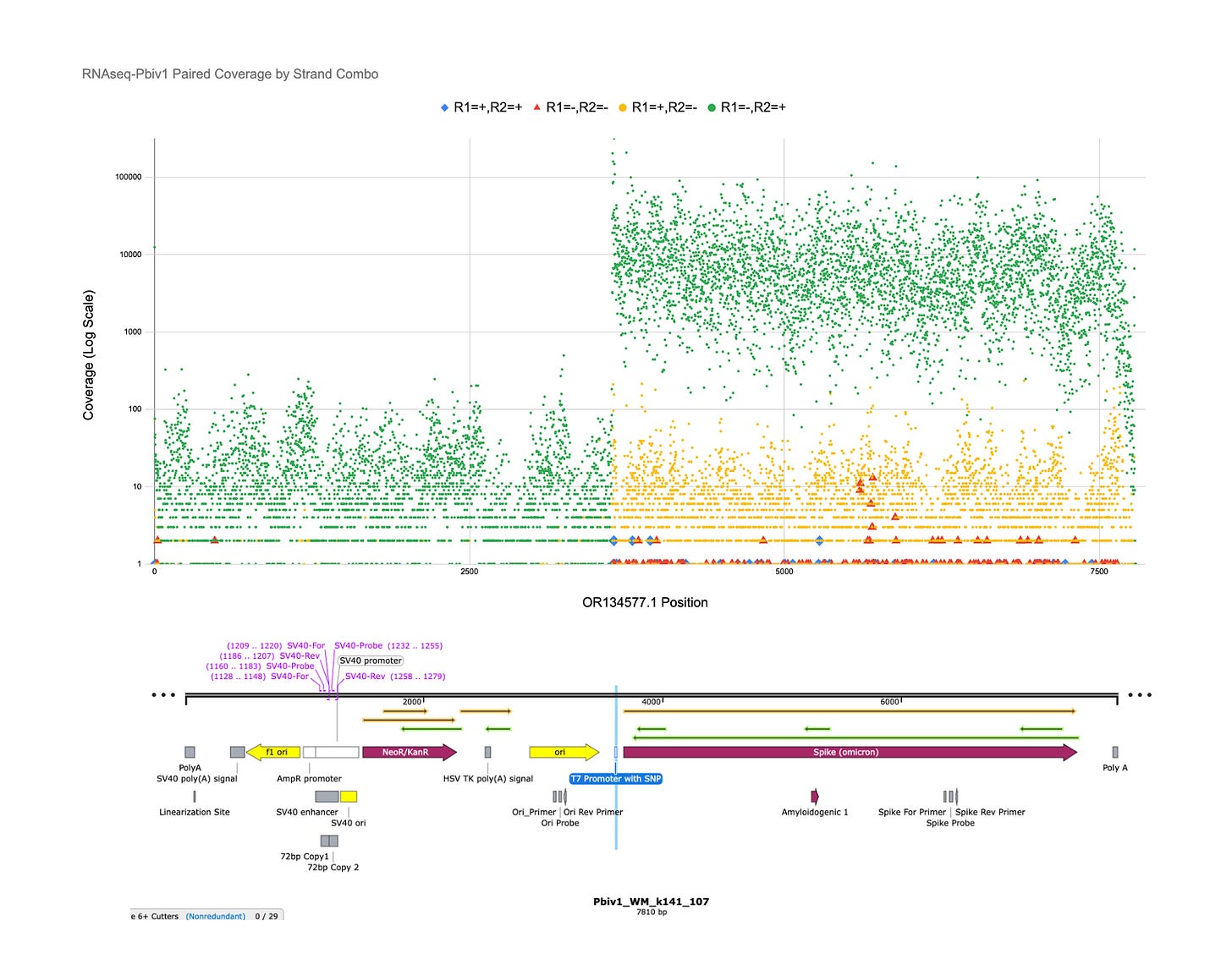
The Crick strands are in Green as R1-/R2+ reads. The Y axis is LOG Sequence coverage and each dot represents one of 7810 bases in the reference plasmid. Most green dots over the Spike region are 10,000X coverage.
The Watson reads are in Yellow as R1+/R2- reads. These could be derived from the plasmid DNA.
The other blue and red reads may be cis or trans template switching with the T7 polymerase manufacturing of the vaccines. These are mostly under 10X coverage or 0.1% of the reads.
Pfizer Bivalent vial 1
Pfizer Bivalent vial 2
All of those yellow reads are Spidroin strand modRNA or plasmid DNA, not Spike strand modRNA. They are 2-3 LOG scales lower in frequency than the Crick strand.
The red and blue dot are locations in the reference where reads that contain paired end sequences that cross strands or orientation. These are likely the template switching prone locations in the plasmid. See 5500-5720bp in the plasmid.
If you look for the secondary structure of this region of the plasmid sequence in OligoAnalyzer from IDT you find many template switching prone palindromic sequences.
Sanity check- Why isn’t there equal watson and crick read coverage over the DNA? Shouldn’t green and yellow be the same? Why is there about 1 LOG more Green than Yellow in the DNA regions of the plasmid?
One would expect there to be equal watson and crick stands in DNA but the use of Actinomycin D in the 1st strand synthesis (RT step) favors the capturing of ssDNA and ssRNA in the vaccine over dsDNA. While there is still less of a gap between yellow and green read types, this is still an anomaly I cant fully explain. The pBR322 origin of replication is a bidirectional origin of replication so I would expect equal reads numbers from both strands unless there is some basal level of plasmid RNA. ONT sequencing has shown higher DNA coverage over the F1 origin which may explain this artifact as this origin is a ssDNA origin of replication and may be active in this cell line?
You can easily see the T7 promoter at base 3621 shoots the crick strand RNA signal up several orders of magnitude.
Does this rare amount of template switching even matter?
Small amounts of dsRNA can trigger the interferon and RNAi pathways. You really want to avoid this. Moderna would not bother to engineer a dual mutant T7 polymerase if this wasn’t a serious concern.
Rare events also matter if we are dealing with amyloidogenic peptides in Spidroin as amyloids can seed an amplifiable cascade in cells.
Template switched mRNAs are unlikely to have a polyA tails suggesting they may get ignored in translation?
They will have all of the correct 5 prime content as they were derived from a normal spike 5’ end. They will get capped and have a Kozak consensus sequence but the 3’ end will be immature.
This is a point of debate. If these mRNAs get to the nucleus (See these papers that suggests it will), they can be endogenously poly-adenylated by TENT5A enzymes. Some poly-adenylation can occur in the cytoplasm with TENT5C as well.
More work needs to be done to assess if these red and blue reads are in fact T7 polymerase errors from the modRNA synthesis or if they are an artifact of us converting RNA into DNA during the sequencing process.
Gholamalipour et al suggest a few ways to tease this apart in RNA-Seq data.
This is still a work in process and there are limitations to this work. There are 2 Reverse Transcriptase reactions occurring. One during the manufacturing and one during the sequencing library generation. Teasing these apart may require UMIs (more DNA barcodes to track the molecules at each step).
At the very least, people should wonder why the dsRNA in these vaccines is monitored with an ELISA assay? Was this assay even functional on N1-methyl-pseudoU RNA? Why not RNA-Seq where fine level detail like this can be seen? This is likely very relevant as the Electropherograms of the mRNA has signs of more dsRNA with process 2 in the form of worse RIN scores.
Stay Tuned…
















Thank you Kevin. I pray that the world sees and understands the massive fraud taking place in real-time. Peace.
For the reverse strand ORF... it would be a substrate for 5' capping, but would lack the Kozak. So it seems chimeras as described above would be more likely to result in translated protein?