Fluorometry Deep Dive
The role for DNaseI and RNaseA
This is an important technical topic as the regulators like the EMA are being handed qPCR data for the DNA contamination and Fluorometry data for the RNA levels so it is important we understand the limitations of these methods.
This thread will serve as a background on these methods. Once you understand the limitations of these fluorescent dyes it will become a lot more apparent to you that Pfizer intentionally deceived the regulators with their use of RiboGreen as a RNA measurement assay.
Most of the fluorometry methods rely on dyes that lack fluorescence until they are bound to DNA or RNA and upon binding to nucleic acids their fluorescence increases 1000 fold. This gives them 3 orders of magnitude of dynamic range. This is lower dynamic range than qPCR which can achieve 6 orders of magnitude in dynamic range.
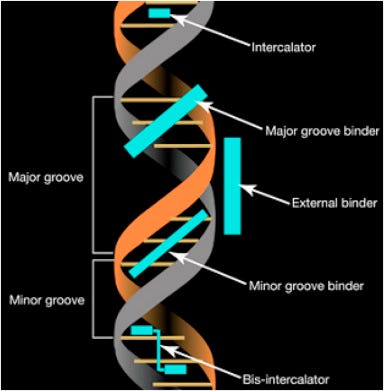
The fluorometry dyes are known as Intercalating dyes. They are minor groove binding dyes which require dsDNA and sometimes dsRNA to bind. They are only fluorescent when bound to a minor groove. Tolun et al. depict PicoGreen binding to dsDNA with Lambda Exonuclease eliminating the fluorescence and monitoring this in realtime.
Above is a depiction of Major and Minor grooves in DNA. PicoGreen and AccuGreen are minor groove binders which have a 4bp binding site. This is going to become important as we look at the minimal size of DNA that can trigger PicoGreen signal.
Image taken from Dragan et al.
Fujihara set out to test this with electrophoresis as the readout.
Note, they are interested in using circulating DNA as a marker for acute myocardial infarction (AMI).
DNaseI activity in the serum is up-regulated within 3 hours in AMI patients. This marker can be detected before troponin levels increase.
What is important to note is that DNaseI doesn’t eliminate all PicoGreen green signal. This is likely due to the fact that DNaseI has a minimal footprint of DNA required for catalysis. It needs to bend the DNA 20 degrees to open up the minor groove for hydrolysis and this is more difficult to do with PolyA tracts that are present in the modRNA plasmids (Rojo et al).
Choi et al is another good read on the topic.
The other potential DNaseI inhibitor to consider is the intercalating dye itself. If this is present in the minor groove, does it inhibit DNaseI from digesting DNA? This is something we set out to test below.
Lysing open the LNPs is an important consideration when measuring with these dyes. We have usually relied on the heating and cooling in qPCR to take care of this.
With fluorometry, one can pre-boil the samples but there is concern that cooling the sample back to 4C or RT may allow some LNPs to reform and protect the nucleic acids from digestion.
Another approach is to use a surfactant. But you need to pick a surfactant that doesn’t inhibit enzyme function. LiDS or SDS are anionic detergents which can inactivate some enzymes. A better choice is a non-ionic detergent like Triton-X that can accelerate the activity of some enzymes.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7416074/
You can see this recommendation in the manufacturers literature. We settled on 0.1% TritonX-100 to break open the LNPs.
Let’s review what we want to do.
We want to quantitate the amount of DNA in a vial where it is swamped with RNA.
The challenge with direct PicoGreen/AccuGreen measurements is that there is some cross talk between DNA and RNA binding with that dye. Any dsRNA will attract PicoGreen.
You can see RNA alone produces small amounts of DNA signal but it is not zero. While the 1:1 DNA:RNA line looks unbiased, you have to consider what happens when the RNA is 100X more than the DNA (as it may be in a modRNA injection). You can get more signal from the RNA than from the DNA, leading to inflated DNA signals.
There is also another dye used by Pfizer known as RiboGreen which is more designed to fluoresce in the presence of RNA but even it has more severe cross talk with DNA. The signal is twice as high with equal amounts of DNA so its suggested to use this with only DNaseI treated samples. This 2:1 ratio of DNA:RNA signal for RiboGreen is better than PicoGreen which has a 100:1 preference for DNA over RNA but it really shouldn’t be called RiboGreen given it binds DNA more favorably than RNA.
Jones et al. describes this in their paper first describing RiboGreen.
Pfizer used this trick to measure the RNA for the regulators. The contaminating DNA is inflating the values of the the RNA as RiboGreen is known to produce 2X more signal on DNA than on RNA (See figure 7 above).
While PicoGreen has the opposite effect. It has~100X more signal on DNA than RNA but this relationship gets messy with DNA-RNA hybrids and RNA with modified bases.
.
So the take home message is that PicoGreen (DNA dye) is more specific for DNA/RNA than the RiboGreen (‘RNA’ Dye but really binds DNA better) that Pfizer is using to measure their RNA. With every DNA contaminant they have, they get 2X more ‘RNA’ signal. They then turn around and measure their DNA with qPCR which under counts their DNA value. It is actually a very clever trick they pulled off once you understand that RiboGreen has 2X more signal on DNA than RNA.
Lets consider the EMA Specification of 330ng/mg of DNA/RNA. You need to hit 1 DNA molecule per 3030 RNA molecules. If you measure the RNA with a tool that double counts RNA for every DNA molecule you have, and then measure the DNA with a tool that Moderna’s own patents declare undercounts the DNA, you can game these regulations very easily.
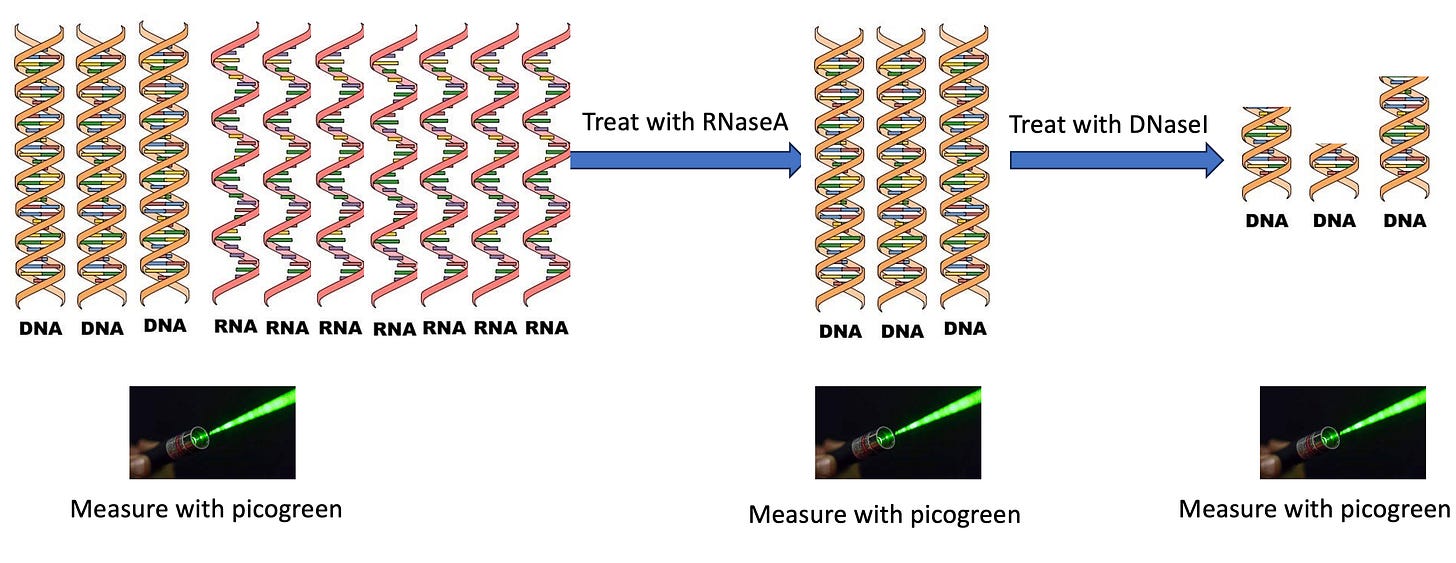
For our application, we are not trying to game a regulation. We are trying to use these Dyes where they have their strongest application justification. So lets use the more specific PicoGreen dye to measure DNA and monitor what happens when we eliminate the RNA with enzymes like RNaseA and DNaseI.
If DNA is the minority species in the vial and we know large amounts of RNA can dominate your DNA signal, we can’t just take the PicoGreen on face value. We mentioned this in Speicher et al. There is a lot of literature on how these dyes behave with dsDNA, ssDNA and RNA but there is zero literature on what modRNA does to these dyes.
This why RNaseA and DNaseI are so important as they allow you to remove one nucleic acid very selectively and remeasure things. This holds true for the majority of the DNA in the vial as long as DNaseI can digest all DNA in the vial. As we can see from Fujihara et al, it removes most of the DNA but it doesn’t digest it all .
RNA vs DNA differentiation is even more problematic for UV spectrophotometry as the RNA levels exceed the DNA levels, cross talk from the UV spectra of RNA bleeds into the DNA signal and you get confused really fast.
So what happens when you take all this advice into consideration and attempt to use DNaseI to digest our fluorometry standards. These have no LNPs interfering with the read out and this should give us a floor of DNA that cannot be digested but is still PicoGreen stainable.
For this experiment we are using a DNaseI from NEB known as DNaseI-XT. This enzyme is optimized to handle DNA generated from an In-Vitro Transcription reaction. The Enzyme is a bit more salt tolerant than DNaseI and is designed to handle DNA:RNA hybrids.
In this experiment, we perform a time course of DNaseI-XT treatment on the Standard curves used to calibrate the Fluorometer.
One can see the DNaseI-XT reaction is complete within 1 hour at 37C with 3ul of the Enzyme (2Units/ul). The reactions were performed with 30ul of Standard DNA, 3ul of Enzyme and 3.7ul of 10X reaction buffer. PicoGreen (AccuGreen TM) was added after this reaction and compared to samples that were further digested for additional time in the presence of PicoGreen. We don’t see PicoGreen inhibiting DNaseI-XT. The same DNA floor is hit at ~100 picograms wether the reaction is performed in the presence of PicoGreen or when PicoGreen is added after the reaction.
So there is some DNA in the standards that remains stainable but undigestible with DNaseI-XT. Nevertheless, DNaseI-XT can reduce the DNA 100 fold in an hour.
This is helpful to know as we want to RNaseA treat the RNA, then measure the DNA and then prove that the remaining signal is in fact DNA that is digestible by DNaseI-XT. We have to just be aware that not all of the DNA will be eliminated.
In this next test we take Pfizer monovalent vaccines and 2 Comirnaty vaccines from Japan and treat them with RNaseA. We do see an order of magnitude drop in the AccuGreen signal. AccuGreen is a competitive intercalating dye to PicoGreen.
We can also see the TritonX addition to the vaccine doubles the signal. Repeated applications of DNaseI-XT doesn’t lower the DNA below the DNaseI-XT treatment of the LNP-free Standard DNAs. This is likely the result of the minimum footprint of the DNaseI-XT. The enzyme can not degrade all of the DNA to an un-stainable size and may have some sequence context limitation in Poly A tracts.
The chart is on a LOG scale. This ranges from 17.5-61.8 ng/dose with the lowest sample being a Comirnaty vaccine from Japan. The DNA remaining after 2 rounds of DNaseI-XT digestion is degraded down to the DNA levels observed with the standard DNA digestions with DNaseI-XT.
This is still an order of magnitude over the qPCR quantitation.
This is inline with a Moderna patent that specifically states that qPCR under estimates the total DNA present in the vaccines.
Conclusions
The use of RiboGreen in the evaluation of the RNA to DNA ratios for injectable products should be banned. This dye provides false RNA values since it more favorably binds to DNA than RNA (Jones et al). Thus any manufacturer looking to improve a RNA/DNA ratio will be motivated to have MORE DNA contamination as the qPCR used to monitor the DNA will respond linearly to the DNA measurement and undercount the small molecules but the RNA measurement will improve by a factor of 2 with the use of RiboGreen on DNA contaminated samples.
Pfizer already had a qPCR assay targeting the spike sequence to monitor their DNA levels. They simply needed to add a 10 minute reverse transcriptase step to provide a RT-qPCR measurement of the RNA. The fact that they intentionally avoided this and moved to a RiboGreen assay known to severely inflate RNA levels when there is DNA contamination is an overt intention to deceive.






















Seems to me that it doesn’t matter what slam dunk evidence is found on the skulduggery of pfizer, the fda, government will do zip about it. Too much corruption, too much money, too many Positioned people with a vested interest. We are on our own. We need a diagnostic test. But, not sure that will even help.
Thank you to all who are on this extraordinary investigation of this enormous crime which killed my 2 sisters our son and my friend