How long are the DNA fragments in BNT162b2?
What can Nanopore sequencing show us
The critics of our work have quickly shifted their arguments from the “DNA does not exist in the vaccines” to an admission that DNA does exist but the length of the DNA that is found is nothing to worry about. Much of this goal post shifting has occurred since Dr. Sin Lee and Dr. Buckhaults have confirmed the presence of the DNA. Dr. Buckhaults performed this confirmation with his own vials that have proven chain of custody. His CT scores are 18-19 while the vials we have in our possession are 15.5-17CT. Now that the presence of the DNA has been confirmed, the arguments have shifted to question its exact quantity (which we expect to vary vial to vial based on the EMA disclosures) and to critique the length of those DNA fragments.
It is true that the Illumina sequencing we first performed on these vaccines has read length limitations that can’t address this question. Some guidelines only concern themselves with DNA that is over 200bp in size. However, the logic behind the fragments under 200bp not being relevant is flawed. They assume DNA under this size range cant code for an Open Reading Frame (ORF) or if it does it will create such a short peptide (66 amino acids) that it wont be relevant or the DNA being short will be broken down quickly.
A few reasons this argument is flawed.
1)The regulations didn’t anticipate this DNA being inside nuclease resistant LNPs, therefore short length doesn’t imply rapid degradation.
2)Even small DNA fragments can be functional. The 72bp SV40 Enhancer is one such element that is known to recruit transcription factors and mobilize the DNA to the nucleus. David Deans work covers this.
3)Even small fragments, if integrated can knock a human gene out of frame and cause problems.
But let us address the problem directly and survey the DNA with sequencers that have unlimited read length.
Oxford Nanopores (ONT) are great at this. While they have longer read lengths, they come with more sequencing error. Additionally some of the ONT techniques selectively clone and sequence shorter fragments so this fragment length assessment will be biased to count smaller molecules more readily than longer ones but it can still provide some information.
From the ONT community page-
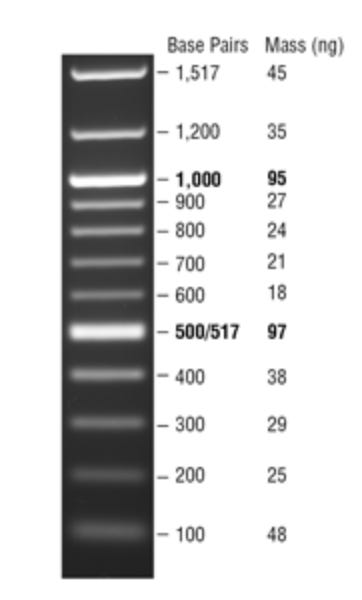
We haven’t had success using the ONT rapid kits mentioned in the above tech-blog. There may be a minimum size fragment these transposon based approaches can work with due to the footprint of the transposase enzyme. Once we migrated to Ligation based kits we began to see more reads but we know these reads are biased for shorter fragments in the library construction steps. There is also a diffusion bias. The shorter fragments diffuse faster and load the pores preferentially over the longer fragments. Ultimately we will need to normalize for these effects by ONT sequencing a DNA ladder with known concentrations of different sizes (Figure 1). That is an experiment for another day.
Until then, what do we see?
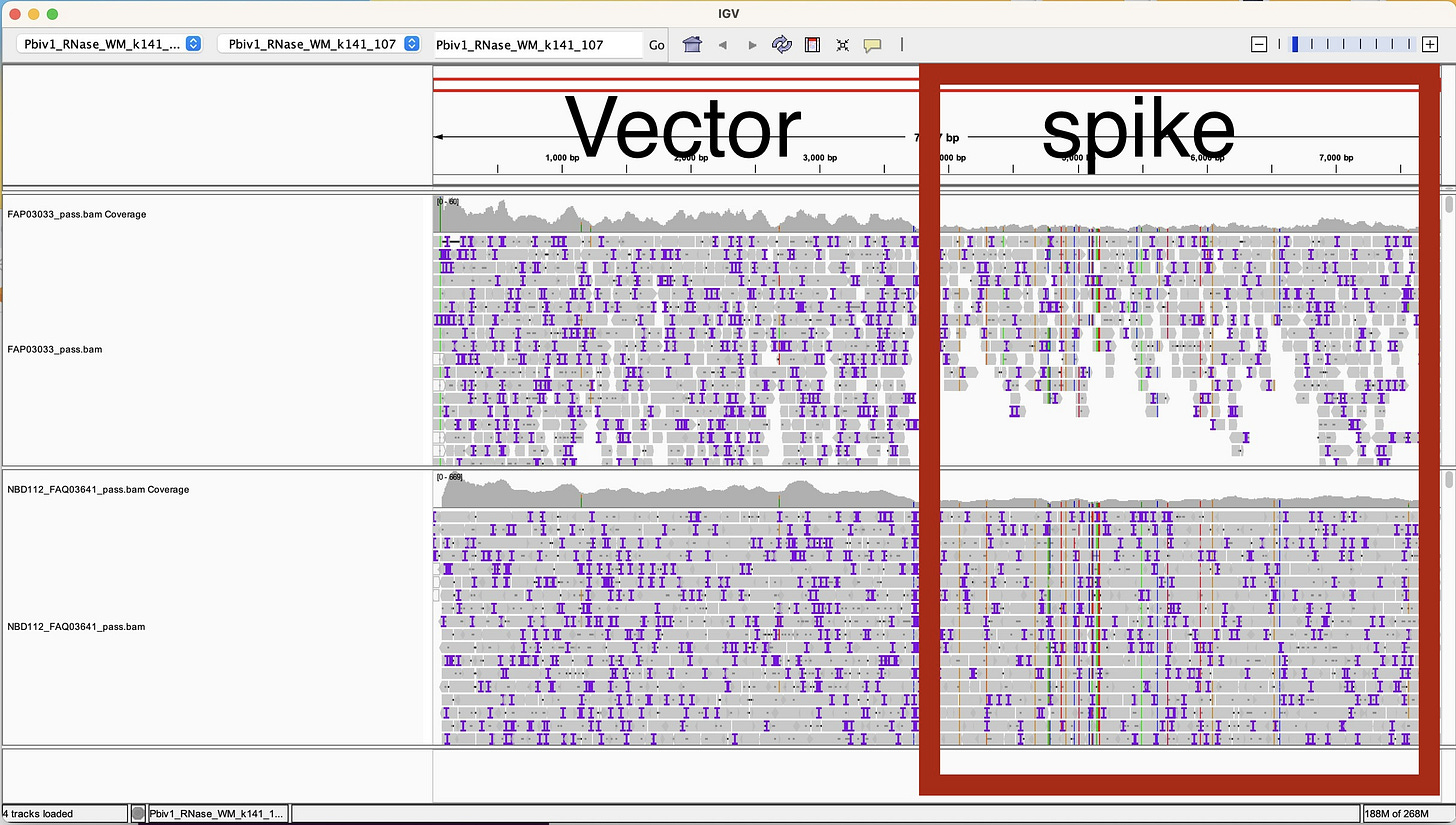
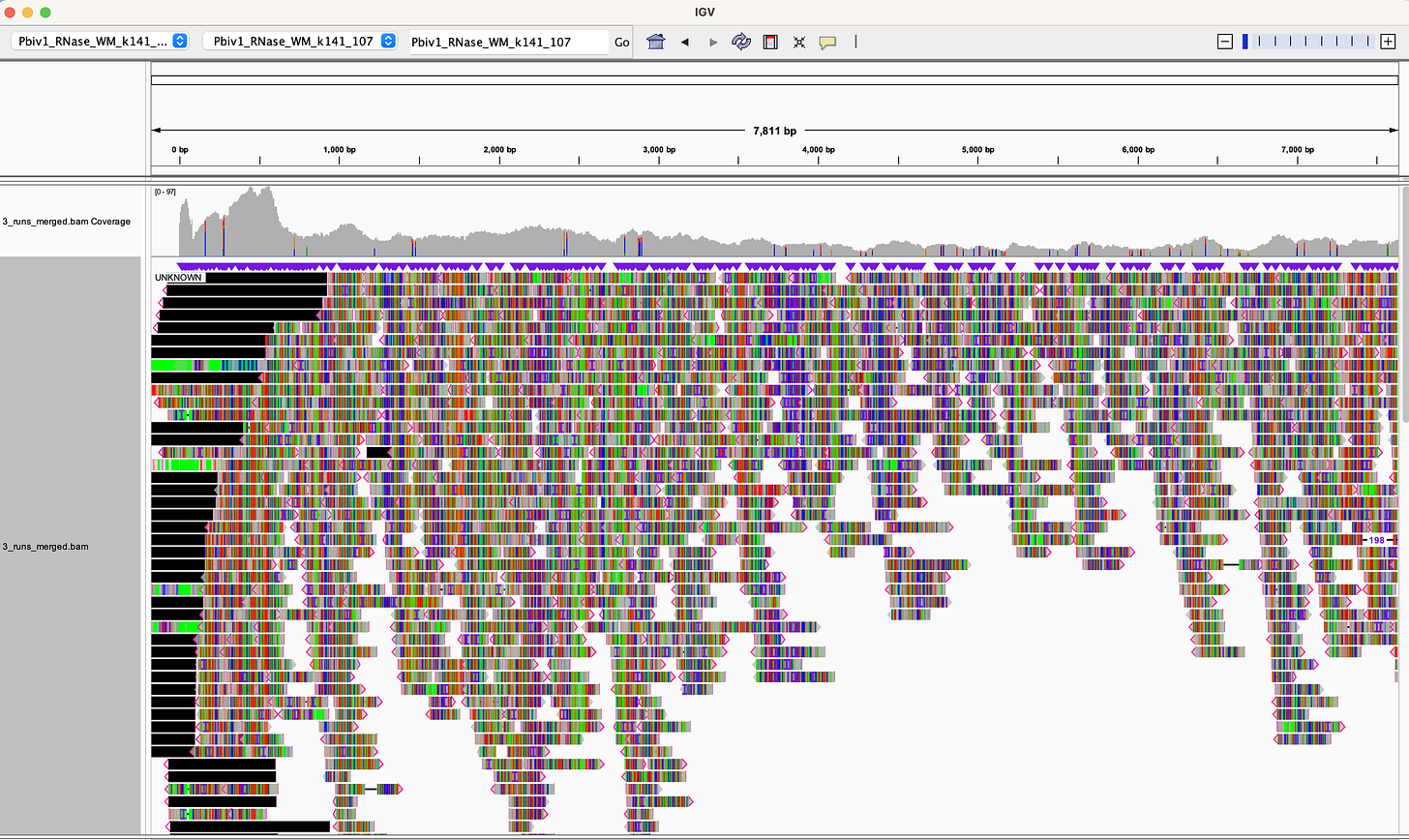
Dr. Buckhaults produced some ONT data on an earlier ONT kit protocol (V9). He purified DNA from 15mls of vaccine. He did not use an RNase A step to get rid of the RNA and while he got great coverage across the entire vaccine plasmid, we can see there is lower sequence coverage over the spike portion of the sequence than the vector (Grey top track in IGV viewer below). The IGV view (Figure 2) was generated by our team from the data Dr. Buckhaults placed public on Twitter.
This difference in coverage over the vector versus the spike region is likely the result of RNA/DNA hybrids that interfere with T4 DNA ligase. These hybrids exist over the part of the vaccine that is turned into mRNA (Spike) but do not exist over the vector sequence. T4 DNA ligase only ligates double stranded DNA so if some of the DNA is hybridized to RNA, those molecules wont ligate with T4 DNA ligase. We used T4 DNA ligase for the SOLiD sequencer so I have a lot of experience with what modified nucleotides it will and will not accept and fully expect there to be a ligation bias with RNA/DNA hybrids.
His data can be downloaded at this link.
We also have data that demonstrates T5 Exo and DNaseI are inhibited by RNA/DNA hybrids so the DNA molecules that are nuclease protected by the RNA are likely to be the longer molecules in the library and thus be under surveyed with a method that doesn’t remove the RNA before ligation.
This led us to attempt a RNase A digestion before Ligation on ONT. We don’t have as many vials as Dr. Buckhaults so we attempted this on 750ul of the Pfizer monovalent vaccines (20 fold less material than Dr. Buckhaults).
We get far fewer reads (866) but they look more evenly distributed. There is some elevated coverage over the PolyA tract as that low complexity region of sequence is recruiting some noisy reads (Black) into the picture.
The mean read length of our data is 214 bases. This is over the 200bp guideline. Most striking in the data is the 2.6Kb-3.5Kb reads that emerged.
A close up of the reads under 1000 bases. This is the Aligned read length so the adaptors and other artifacts are not inflating the length of the reads.
Some may discount these reads claiming half of them, if integrated, will integrate in the reverse orientation in the genome and the reverse sequence is likely to be gibberish and not code for spike.
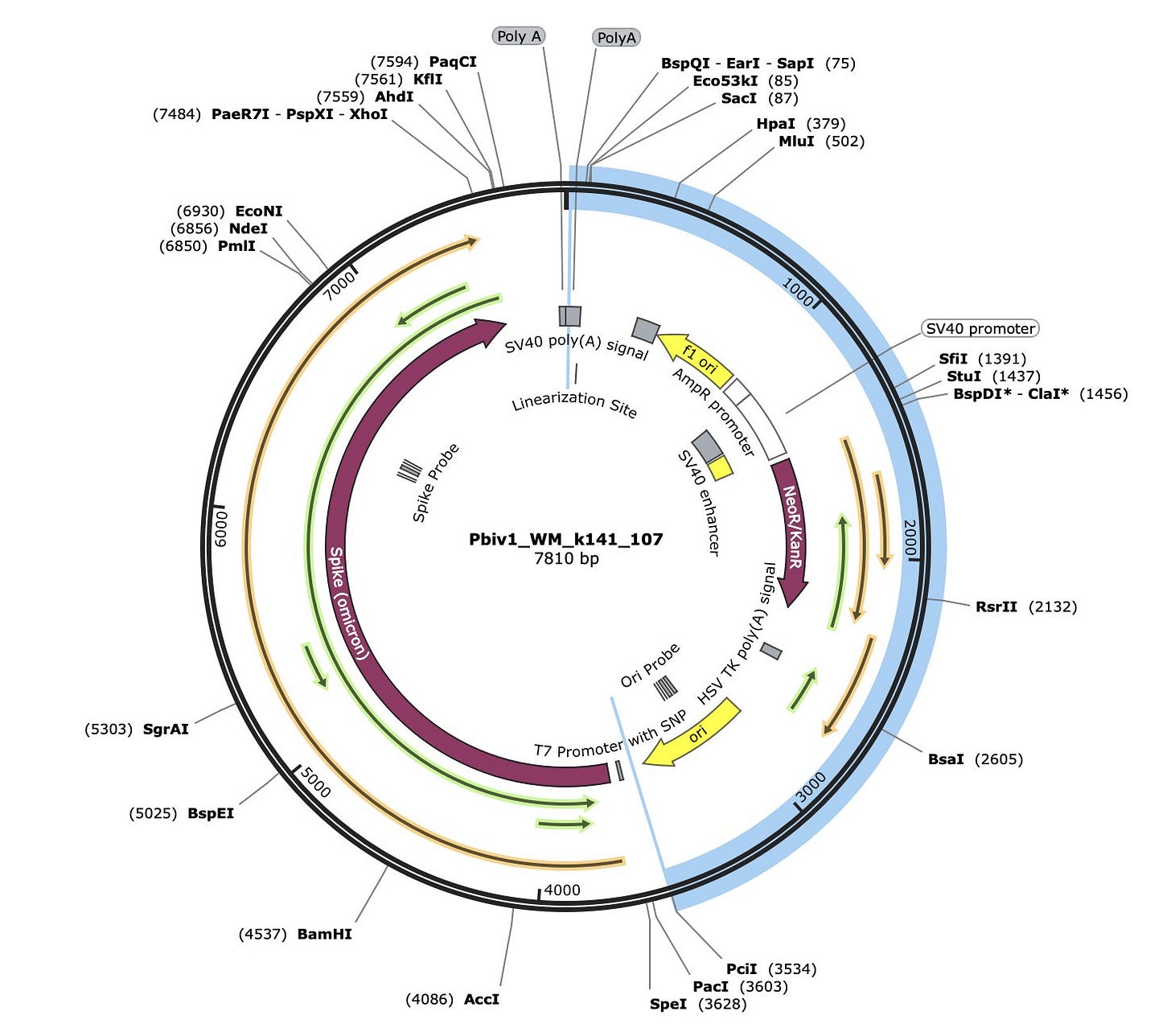
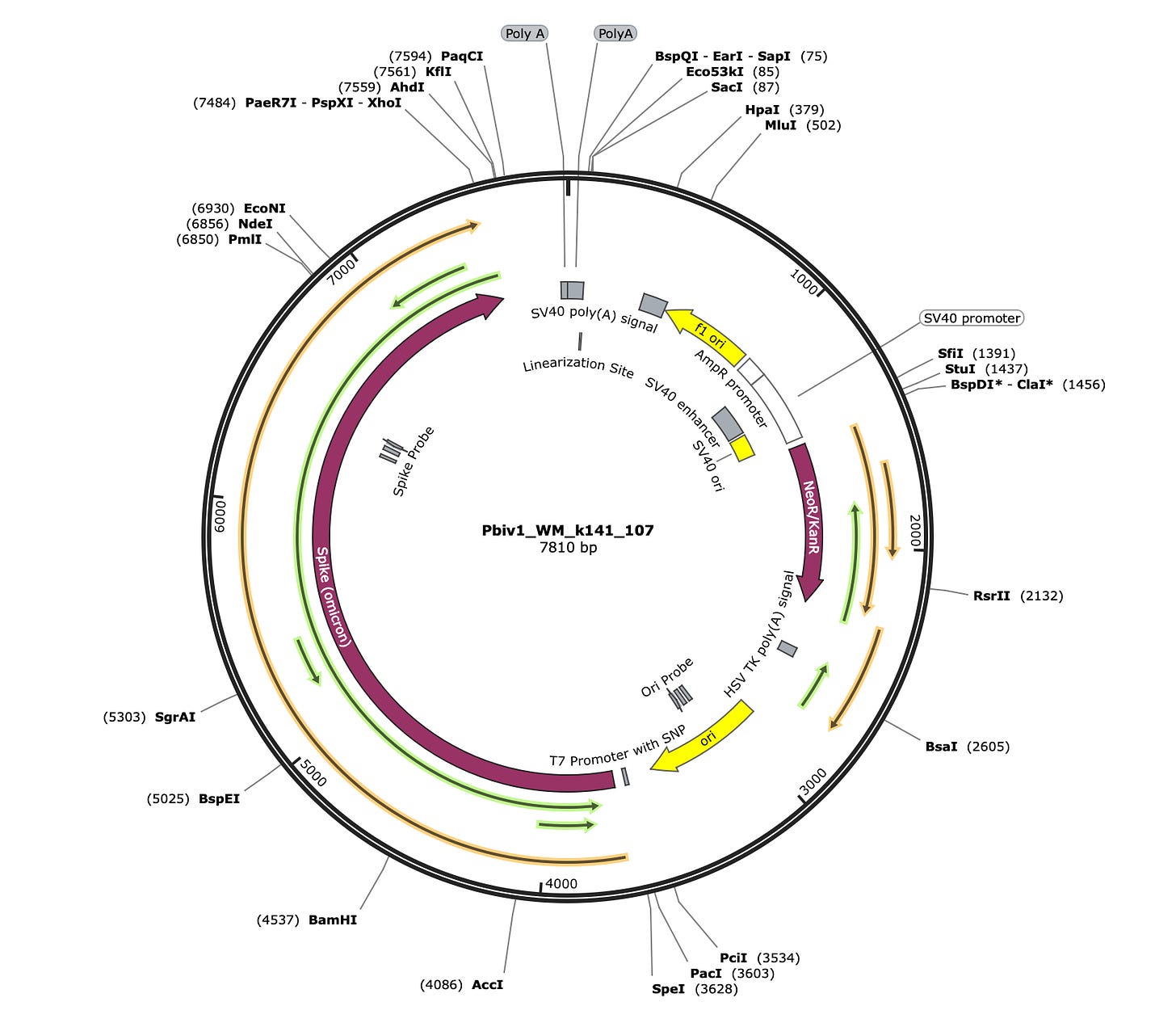
What is surprising to everyone who inspects the Pfizer plasmid is that the ORF that contains the spike protein also has a 1252 amino acid ORF on the reverse strand.
The Orange ORF (Figure 8) is the Spike protein ORF. What is shocking is that the Green ORF pointing in the opposite direction on the reverse strand even exists. This is a remarkable codon optimization feat and arguably a codon optimization failure.
You do not want long reverse strand ORFs like this. If your T7 Transcription of the mRNA from the plasmid ever strand flips (Template Switching of T7 Polymerase) on you, the polymerase can continue to make mRNA in the opposite direction that has no stop codon for long stretches of sequence. This could explain the larger than anticipated molecular weights of spike expressed from the vaccine seen on the Western Blots (See BlotGate by Jessica Rose and Jikkyleaks).
In summary, long DNA fragments are found in some vials. We know the EMA has documented a 815X variance in dsDNA contamination across 10 lots from Pfizer and made note of a poorly characterized DNase assay. While neither Dr. Buckhaults or our lab has been able to reproduce plasmid transformation in E.coli, its possible lots exist with poorer performing DNaseI treatments (seen by the EMA) and in fact have full length plasmids (7810bp). More lots need to be surveyed to rule this out but the mere presence of 3.5Kb molecules in the one vial we examined suggests full length plasmids may be possible.
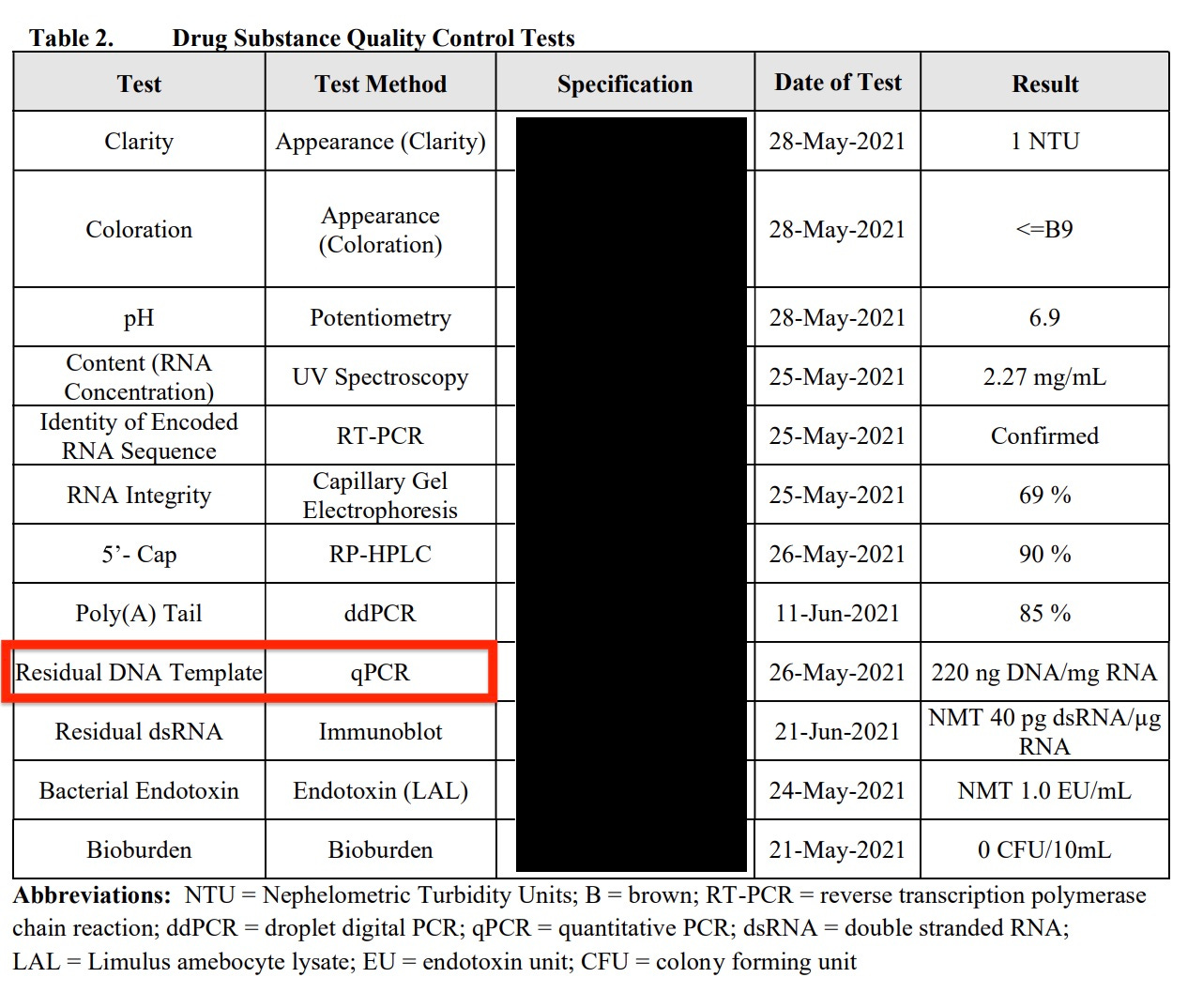
Another detail was brought to our attention regarding how Pfizer is measuring their DNA/RNA ratio. You can see Pfizer is playing games with the regulators.
They use Fluorometry to measure the RNA levels and qPCR to measure the DNA levels!
If you recall, Fluorometry like Qubit and Agilent is what led us to believe 30% of the nucleic acids were DNA, while qPCR gave us much more conservative numbers likely because qPCR is very specific to certain sequences and doesn’t conflate the DNA and RNA signals. These Fluorometry techniques over estimate the nucleic acids while qPCR underestimates the total nucleic acids present but does not conflate DNA and RNA signals.
So why did Pfizer use Fluorometry to measure the RNA and qPCR to measure the DNA when the obvious and more controlled method would be to use qPCR to measure both? This is how they get all their vials to pass testing with regulators. They cherry pick different techniques to cook the books while never being held to disclose the details of each of these detection techniques.
This is often cited as proof that the vaccines are being tested. I can’t blame an accountant for not understanding what a sham this is but they confidently point to redacted reports where the RNA and DNA are measured using different techniques to cook the books. One technique is used to inflate the RNA value and another technique used to depress the DNA number. They should both be measured with qPCR/RT-qPCR so the ratiometric number is normalized to the same technique.
Page 6 of the redacted ‘QC’ for a Pfizer vial.

Appendix-
Brief Methods
V14 ONT Ligation Sequencing kits and V14 ONT Flongles were used to generate this sequence. RNase A treatment was performed according to NEBs instructions and purified with 2X Ampure to ensure small fragments were captured. Dr. Buckhault use different ONT kits and altered the Ligation times to afford better read yields.
ONT sequence for the long reads.
The two long reads aligned above
>Read 1
CTTGGAAAGCTAGCAAAAAAAAAAAAAAAAAAAAAAGCATATGACTAAAAAAAAAAAAAAAAAAAAAAGAAGAGCTCCAACCGGTGTGGTAGCTCCGCCTAGTTCAACATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGATATTGAATTTTCTTCCTAGTGTATAATGTGTTAAACTACTGATTCTTAATTGTTTGTGTATTTTAGATTCACAGTCCCAAGGCTCATTTCAGGCCCCTCAGTCCTCACAGTCTGTTCATGATTGTAATTAGCTATACCACATTTGTAGAGGTTTTACTTGCTTTAAAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCAATTGTTGTTGTTAACTTGTTTATTGCAGCTTAGAACGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTAACGCGTAAATTGTAAGCGTTAATATTTTGTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAAGCTGCAATGCCACAATCAAAAACATATGGTTCAAAAGGGTCGAGTGTTGTTCCAGTTTCAAGACAGTCTCAGTTATCAAGCGAGGCACTGAACGTCAAGGCGAATGTCATCAGGGCCATGGCCCACTACGTGAACCATCACCCTAATCAAGTTTTTTGGGGTCTCAGGTGCCGACAAGCATGAAATTGGAACCCTGAAGGCAGTCCCCATTCAGCTCTACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAAGGAAGAAAGCGAAGGAGTGGGCGCTAGGGCGCTGGCAAGTGTAGCGGTCACGCTGCGCGTAACCACCACACCCGCCGCGCTTAATGCGCCGCTACAGGGCGCGTCAGGTGGCACTTTTCGGGGAAATGTGCGCGGAACCCCTATTTGTTTATTTTTCTAAATACATTCAAATATGTATCCGCTCTATGGTAACATAACTCGATAAATGCTTCAATAATATTGAAAAAAAGGAAGAATCCTGAGGGTGGAAAGAACCAGCTGTGGAATGTGTGTCAGTTAGGGTGTGGAAAGTCCCCAGGAGCTCCGAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGAAACCAGGTGTGGAATTAAAGTGCTCTCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCAACTCTGCCACTCTCCGCCCATTCTCCGCCCCACGGCCATGAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCGGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAGATTGATTAAGAGACAGGATGAGGATTGTTTCGCATGATTGAACAAGATGGATTGCACGCAGGTTCTCTCTGGCTGCTTGGGTGGAGAGGCTATTCGGCTATGACTGGGCACAACAGACAATCGGCTGCTCTGATGCCGCCGTGTTCCGGGGCCGTCAGCGCAGGGGCGCCCGGTTCTTTTGTCAAGACCGACCTGTCCGGTGCCGACGAACTGCAAGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGACGGCGTTCCTTGCGCAGCTGTGCTCGACGTCGTCACTAAGCGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGCAGGATTGCCTCATCTCACCTTGCTCCCGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCTACCTGCCCATTCGACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATCTAAGCCGGTCTTGTCGATCAGGATGATTTCTGGACGAAGAACATCAAGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTGAAGGTGAGCATGCCCGACGGCGAAGGATCGTCGTGACCCATGGCGACGCCCGCTCGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGCAACCGCTATCAGGACATAGCGTTGCATACCGTGATATTGCTGAAGAACTTGGCGGCGAATGGGCTGACCGCTTCCTCGTCTTCACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGAGCGGGCACTTTGGGGCTTTAAATGACCGACCAGCGACGCCCAACCTGCCATCACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTCGGAATCGTTTTCCGGGATGCCGGCCTGATCATCCTCCAGCGCGGGGATCTCATGCTGGAGTTCTTCGCCCACCCTAAGGGAGGTGAACGAAACACTGGAGGAGACAATACCGGAAGGAACCCGCGCTATGACGGCAATAAAAGAGAGAGACGCACGGTGTTGGGTTGTTTGTTCATAAACGCGGGTTTGTCGCGATGCTGGCACTTCTGTCGATACCCCACCGAGACCCCATTGGGCGCAACACGCCGTGTTTTCTTCCTTTTCCCACCCCACCCCCCAAGTTCGGGTGAAGGCCCAGCGGCTTGCAGCCACGTCTGGGCGGCAGGCCCTGCCATAGCCTCAGGTTACTCATATATACTTTAGATTGATTTAAAACTTCATTCTTAATTTAAAAGGATCTAGGTGAAGATCCTTTGATAATCTCATTACCAAAATCCCTTAACGTGAGTTTTCGTTCCACTGAGCGTCATACCGGAGCAAATATTTAAAGGATTCTCTTGATATTCTTTTTCTTTCTTGCGCGTAATCTGCTGCTTGCAAACAAAAAAACCACCGCTACCAGCGGTGGTTTGTTTGCCGGATCGGCAGAATGTACCAAACTCTTTTTCCCAAGGTAACTGGCTTCAGCAGAGCGCAGATACCAAATACTGTTCTTCTAGTGTAGCCGTAGTTAGGCCACCACTTCAATACTTGTAGCACCGCCTACATACCTCGCTCTGCTAATCCGTTACCAGTGGCTGCTGCCAGTGGCGATAAGTCGTGTCTTACCGGGTTGGGACGGGACAAAGTACTATAGTTACCGGATAAGGCGCAGCGGTCGGGCTGAACGGGGGGTTCGTGCACACAGCCCAGCTTGTAGTGAACGACCTACACCGAACTGAATATACCTACAGCGTGAGCTATGAGAAAGCGCCACGCTTCCCGAAGGGAGAAAGGCGGACAGGTATCCGGTAAGCGGGCAGGGTCGGAACAGGAGAGCGCACGAGGGAGCTTCCAGGGAAACGCCGGAAATTTATAGTCCTGTCGGGTTTCGCCACCTCTGACTTGAGCGTCGATTTTTGTGATGCTCGTCAAGGGGGGCGGAGCCTATGGAAATGTGAGCAACGCGGCCTTTTTACGGTTCCTGGCCTTTCGCTTTCAGATATGAACTGACGAAGAACAAGGACAATA
>Read 2
CATATGACTAAAAAAAAAAAAAAAAAAAAAAAAAAAAGAAGAGCTCCAACCGGTGTGGTAGCTCCGCCGTTTAACATCGCCCTTCCCAACAGTTGCGCAGCCTGAATGGCGAATGGATATTGAATTTTTAAGTGTATAATGTGTTAAACTACTGATTCTAATTGTTTGTGTATTTTAGATTCACAGTCCCAAGGCTCATTTCAGGCCCCTCAGTCCTCACAGTCTGTTCACTGATCATAATCAGCCATACCACATTTGTAGAGGTTTTACTTGCTTTAAAAAACCTCCCACACCTCCCCCTGAACCTGAAACATAAAATGAATGCAATTGTTGTTGTTAACTTGTTTATTGCAGCTTATAATGGTTACAAATAAAGCAATAGCATCACAAATTTCACAAATAAAGCATTTTTTTCACTGCATTCTAGTTGTGGTTTGTCCAAACTCATCAATGTATCTTAACGCGTAAATTGTAAGCGTTAATATTTTGTTAAAATTCGCGTTAAATTTTTGTTAAATCAGCTCATTTTTTAACCAATAGGCCGAAATCGGCAAAATCCCTTATAAATCAAAAGAATAGACCGAGATAGGGTTGAGTGTTGTTCCAAGTTCAAGAGAGTCCACTATTAAAGAACGTGGACTCCAACGTCAAAGGGCGAAAAACCGCTGACATTTAGGGCCATGGCCCACTACGTGAACCATCACCCTAATCAAGTTTTTTGGGTCGAGGTGCCCTGCAAAGCACTAAATCGGAACCCTAAGGGAGCCCCCGATTTAGAGCTTGACGGGGAAAGCCGGCGAACGTGGCGAGAAAGGAAGGGAAGAAAGCGAAAGGAGCGGGCGCTAGGGCGCTGGCAGTGTAGCGGTCACGCTGCGCGTAACCACCACACCCGCCGCGCTTAATGCGCCGCTACAGGGCGCGTCAGGTGGCACTTTTGGGGAAATGTGCGGAACCCCTATTTGTTTATTTTTCTAAAATACATTGAAATATGTATCCGCTCATGAGAGATAACCTGAGTAAAAATGCTTCAATAATATTGAAAAAGGAAGAATCCTGAGGCGGAAAGAACCAGCTGTGGAATGTGTGTCAGTTAGGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAAAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTTATTTATGCAGAGGCCGAGGCTGCCTCGGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTTTTTTGGAGGCGGCGCCTTCTGGAACAATTGATCAGAGGTACAGGATGAGGATCGTTTCGCATGATTGAACAAGATGGATTGCACGCAGGTTCTCGCTGTCTTGGTGGAGAGGCTATTCGGCTATGACTGGACAAGGATAGACAATCGGCTGTCTTGATGCCGCCGTGTTCCGGCTGTCAGCGCAGGGGCGCCCGGTTCTTTTTGTCAAGACCGACCTGTCCGGTGCCCTGAATCAACTGCAAGACGAGGCAGCGCGGCTATCGTGGCTGGCCACGATGGGCGTTCCTTGCGCAGCTGTGCTCGACGTTGTCACTGAAGCGGGAAGGGACTGGCTGCTATTGGGCGAAGTGCCGGGGCAGGATCTCCTGTCATCTCACCTTGCTCCTGCCGAGAAAGTATCCATCATGGCTGATGCAATGCGGCGGCTGCATACGCTTGATCCGGCGACCTGCCCATTCCACCACCAAGCGAAACATCGCATCGAGCGAGCACGTACTCGGATGAAGCTGGTTTGTCGATCAGGATGATCTGGACGAAGAACATCAGGGGCTCGCGCCAGCCGAACTGTTCGCCAGGCTCAAGGCGAGCATGCCCTACTGCGAGGATCTCGTCGTGACCCATGGCGATGCCTGCTTGCCGAATATCATGGTGGAAAATGGCCGCTTTTCTGGATTCATCGACTGTGGCCGGCTGGGTGTGGCGGACCGCTATCAGGACATAGCGTTGGCTACCCGTGATATTGCTGAAGAACTTGGCGGCGAATGGGCTGACCGCTTCCTCGTGCTTTACGGTATCGCCGCTCCCGATTCGCAGCGCATCGCCTTCTATCGCCTTCTTGACGAGTTCTTCTGAGCGGGACTCTGGGGTTCGAAATGACCGACCAAGCGACGCCCAACCTGCCATCACGAGATTTCGATTCCACCGCCGCCTTCTATGAAAGGTTGGGCTTCGGAATCGTTTTCCGGCACGCTGGCTGGATGATCTCTCAGCGCGGGGATCTCATGCTGGAGTTCTTCGCCGACCCTAGGGGAGAGGCGAACCTAAACACGGAAGGAGACAATACCGGAAGGAACCCGCGCTATGACGGCAATAAAAAGACAGAATAAAACGCACGGTGTTGGGTCGTTTGTTCATAAACGCGGGGTTCGGTCCCAGGGCTGGCACTCTGTCGATACCCCACCGAACACCCATTGGGGCCAATACGCCCGCGTTTCTTCCTTTTCCCCACCCCACCCCCCAAGTTCGGGTGAAGGCCCAGGGCTCGCAGCCAACGTCGGCGGCAGGCCCTGCCATAGCCTCAGGTTACTCATATATACTTTAGATTGATTTAAAACTTCATTTTTAAACAATACGTAACTGAACGAGTAGATAAACATAAAAAAAAAAA








thank you Kevin,
the medicines regulators reading this know you are right, but they and Pfizer and Moderna are banking on no Court on the planet being able to understand it
but at the end of the day contamination means just that, where gene technology and GMO legislation has been clear for a long time - you are not allowed to threaten, let alone actually poison and dysregulate, any one person's natural DNA integrity
for now Pfizer and Moderna's silent response is clear: 'Mutants!?!? .. Genetic Disorders!?!? .. hey bro, bring them on .. they are good for business'
Why haven't the CDC and the FDA disclosed these issues?
What else are we going to find out years later as independent labs investigate these issues?
I am glad I did not get the #ClotShot.