Lariats and RNA Splicing
Why RNA qPCR tests are inherently different than DNA qPCR tests
Early in the pandemic many people noticed CDC C19 qPCR primers that hit Human DNA. Debunkers quickly reminded people that in order for PCR to amplify, you must have all 3 primers land in proximity (100-400bp) to one another. So single primer alignments to different distant locations in the genome could be ignored.
Fact Check: True for DNA. False for RNA.
Here is the Nuance-
Let’s take the US CDC primer sequences and BLAST them against the Human genome.
You’ll get many hits but you need to inspect them to see if they include the 3’ end (right side) of the primer in the alignment as the 3’ end is the business end of the primer as the polymerases interrogate that region with high fidelity.
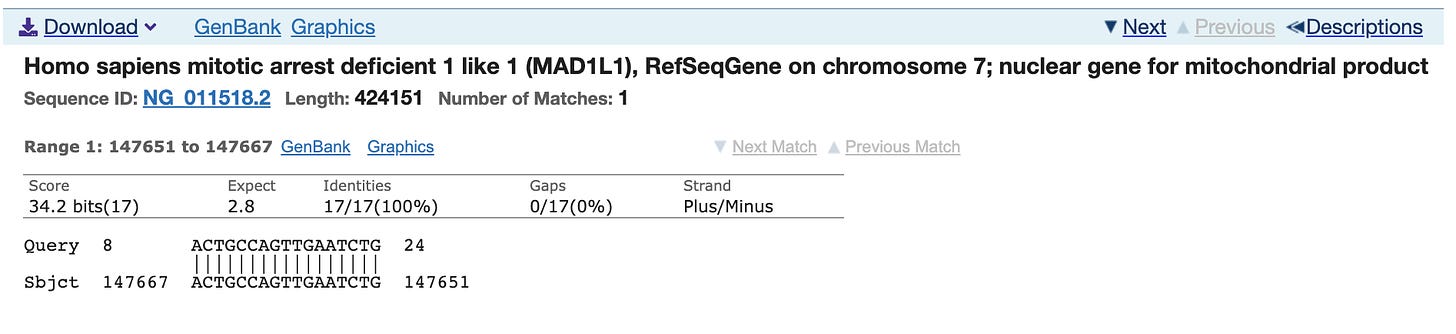
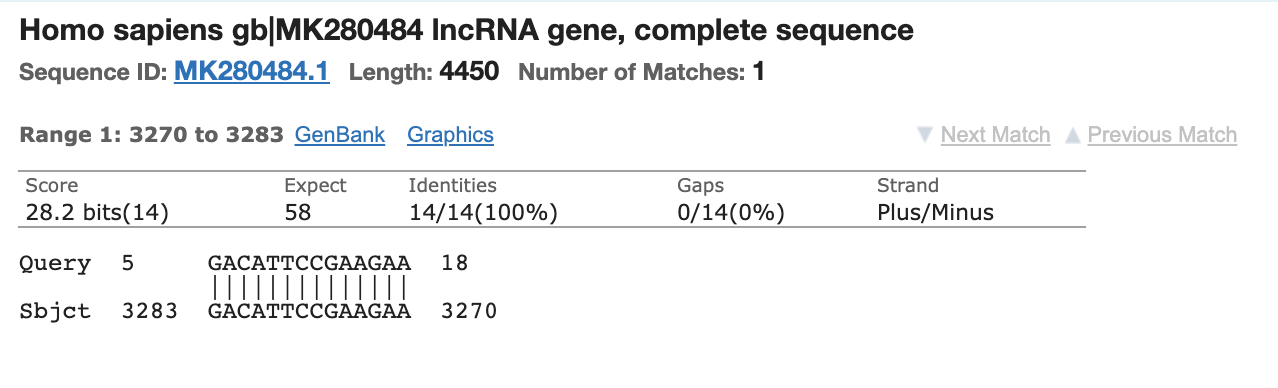
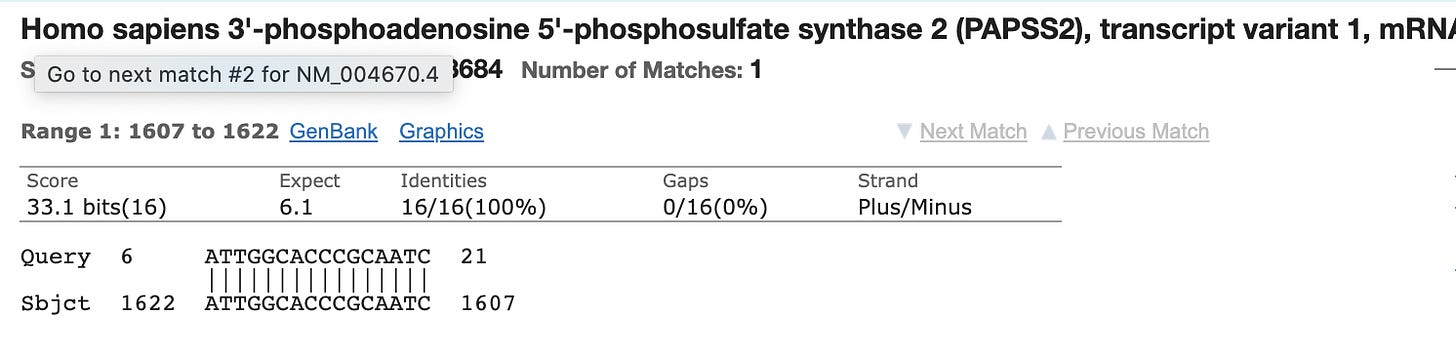
This is the Reverse primer for the N1 primer pair.
It’s never good to see Mitochondrial hits as those sequences are at 500-1000X higher copy number in the cell than the nuclear genome. This happens to be a nuclear Mitochondrial gene (few). Yes, over 1200 genes in the nuclear genome have mito localization signals.
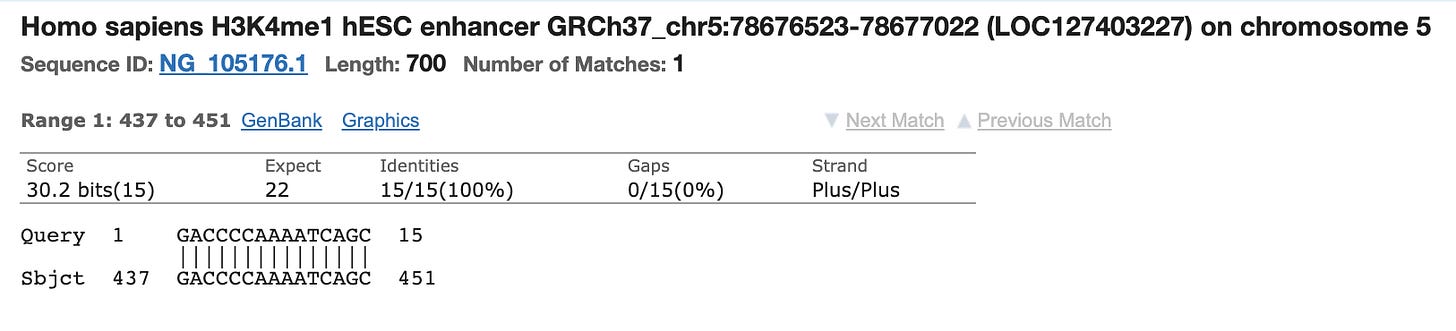
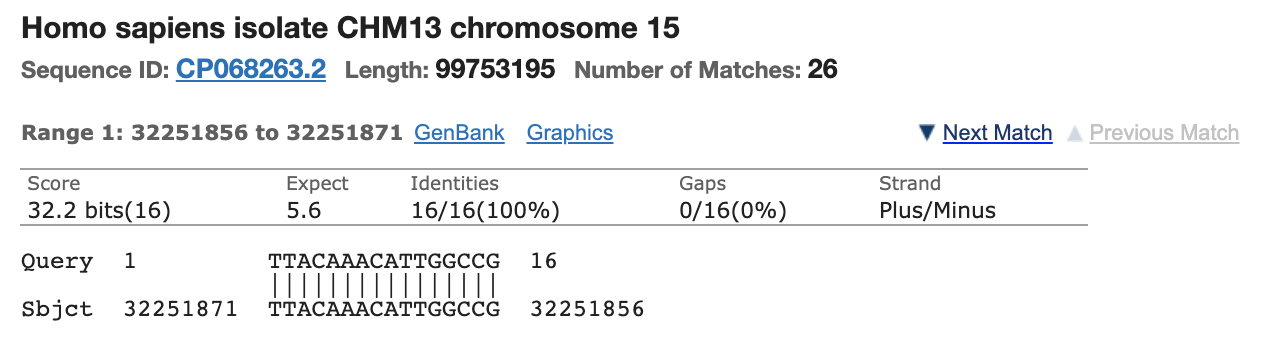
Most people discounted this finding as the other primer only has homology to Chromosome 5 and its homology is only 15 bases and it doesn’t include the AAAT sequence on the 3’ end of the primer. You can’t get a PCR product to form between different chromosomes in the genome as the target DNA is not contiguous.
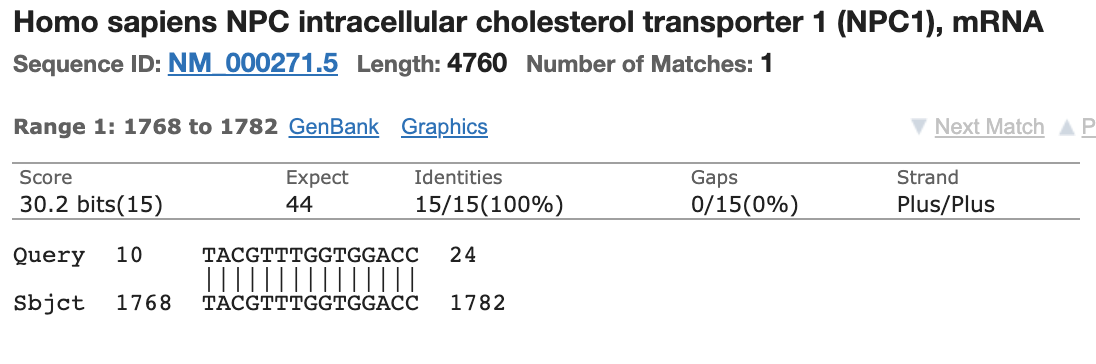
The probe for this N1 probe set also has partial (14bp/24bp) homology to an Human mRNA.
This is not ideal but not lethal.
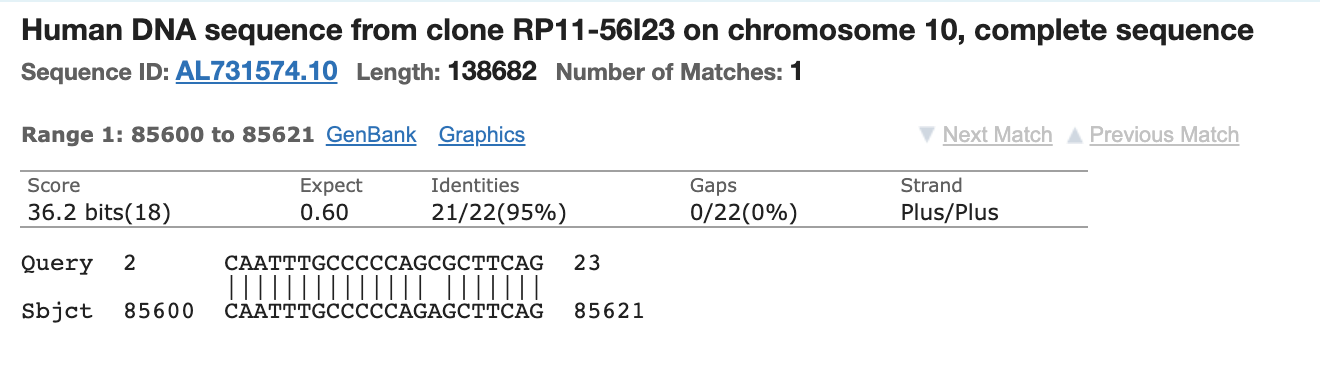
The N2 primer set also has partial matches.
N2 Forward
N2 Reverse has 3 prime homology but is 5 bases short on the 5’ end.
N2 Probe has 21/23 bases matching.
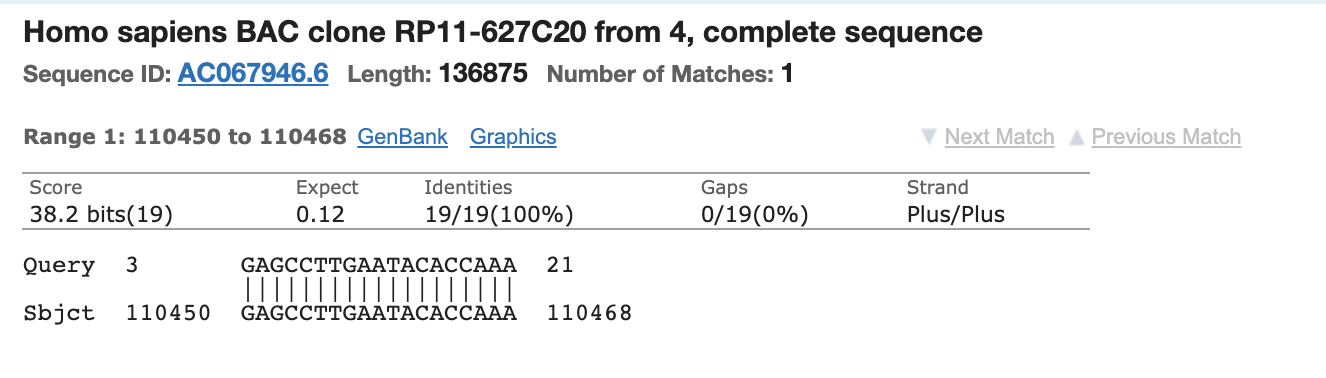
N3 Forward has 3’ homology
N3 Reverse is missing 1 base on its 3’ end.
N3 Probe has only the middle part of the probe with homology.
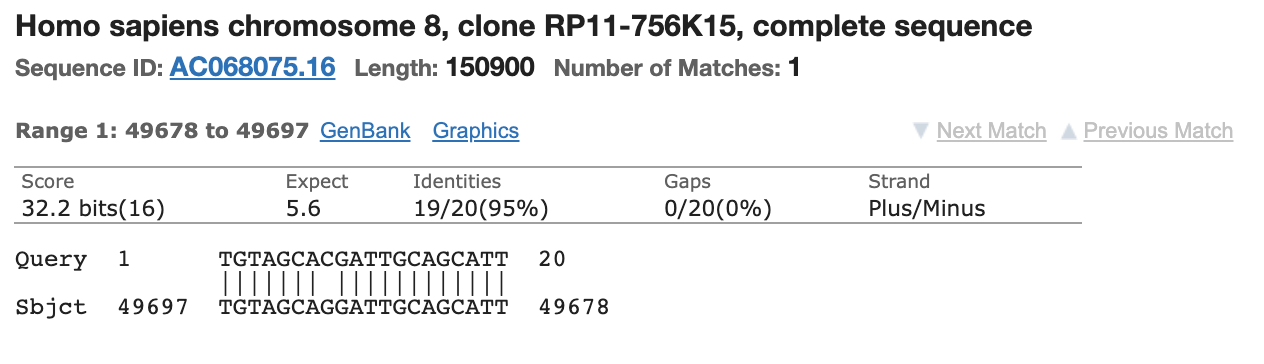
This didn’t just impact the CDC primers. This was highlighted in the Corman-Drosten Review addendum where one RdRp primer had 3 prime homology to Human chromosome 18 but was quickly excused as the other primer were clean.
What does one make of this?
Well, the FDA, in its glorious PCR wisdom put together some guidelines for the EUA qPCR tests that some people seem to be abusing. They have brought out the Holy Hand Grenade and declared that in-silico screens like this should not have more than 80% homology to other background flora (ahem… human is background here). When you see EUA, you should think quick and dirty Emergency generated kits with a get out of jail free card.
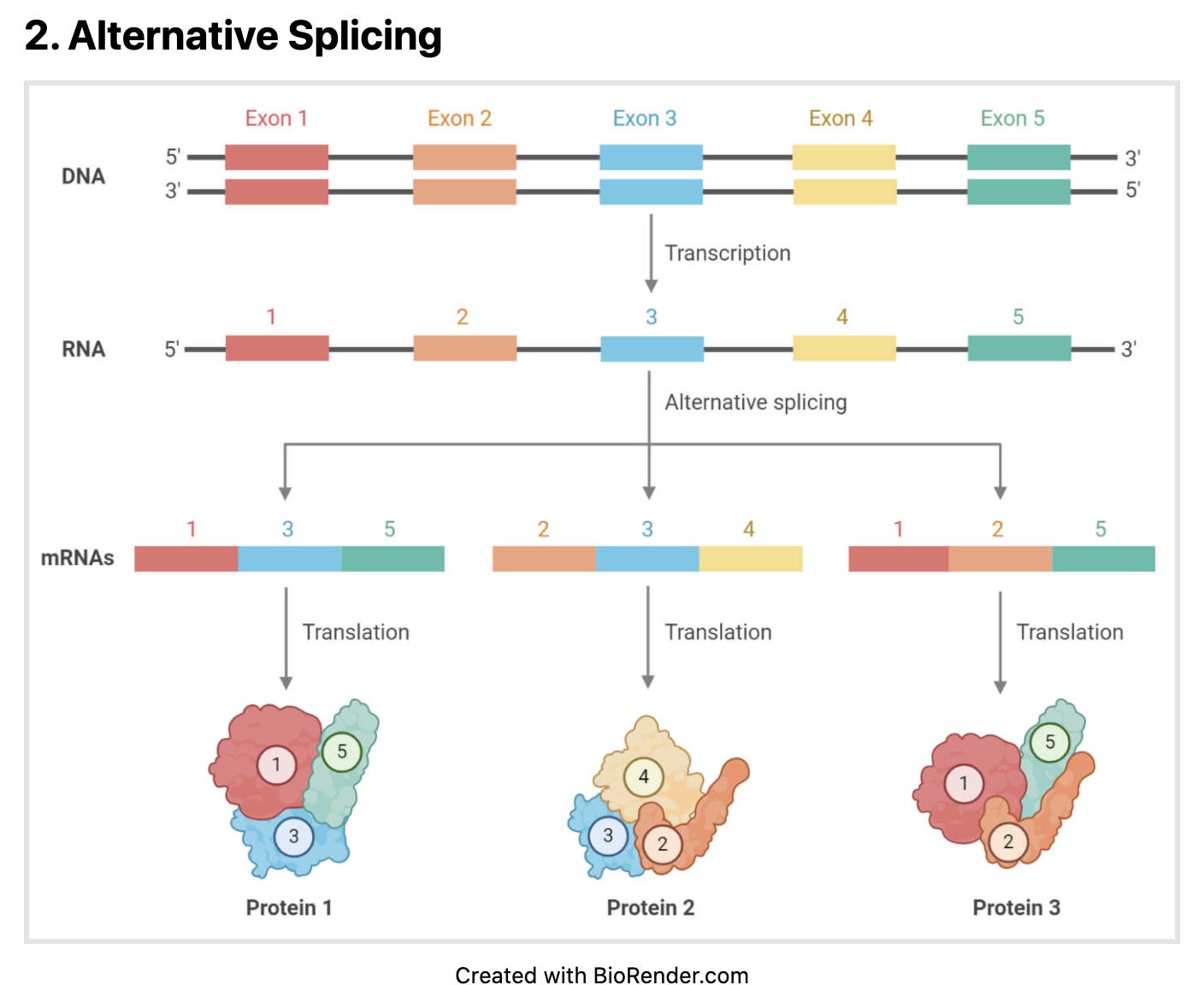
This problem gets more complicated with RNA based tests as the splicing machinery shuffles the genome while turning it into mRNA. So now your primers that were once believed to be too far away on the genomic coordinates to amplify, are now in close proximity.
You are probably not going to see primers on different chromosomes splice together into an mRNA but trans-splicing isn’t impossible.
So if your test is targeting RNA and DNA (which all RT-qPCR kits do by default), then you really need to perform a much more permissive in silico search for potential primer target coordinates.
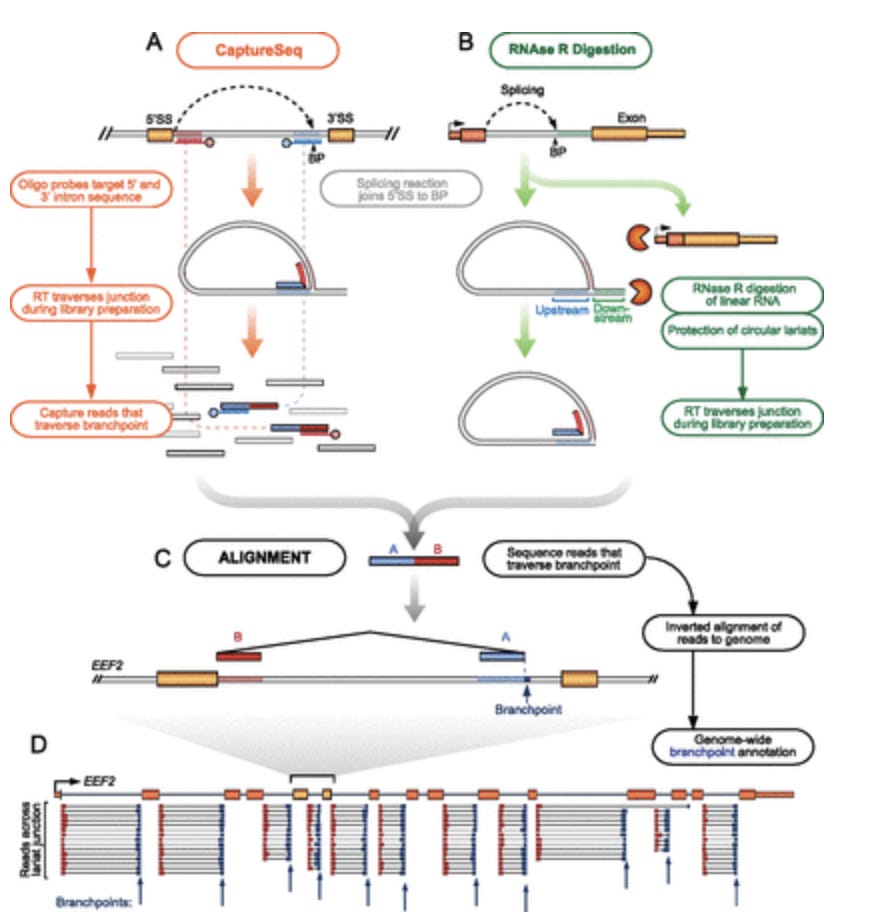
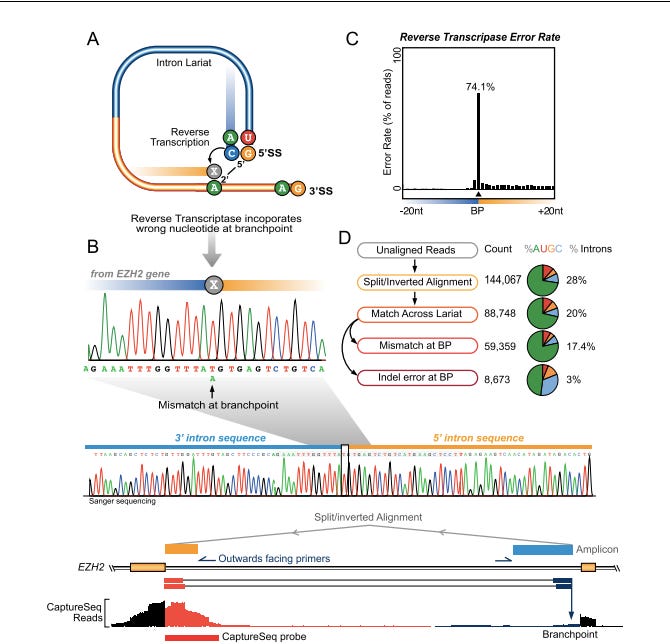
The problem gets more complicated once you learn about Lariats. These are circular RNAs that are formed by Back Splicing introns. This paper finds 59K of them in the transcriptome of a given cell line. These are sequences that are now inverted in orientation through circularization so your assumptions about primer orientation and distances is once again fooled by mother nature.
They develop some clever techniques to measure these with NextGen sequencing.
So once again, when you are designing RT-qPCR assays that target RNA, you need to consider orders of magnitude more complexity than just a genomic DNA reference.
For this reason, its really important that you don’t just stop at an FDA 80% in silico screen of your primers. You really need to test them on live organisms with active transcriptomes to measure off target effects.
Some assays on the C19 PCR market did this. Many did not.
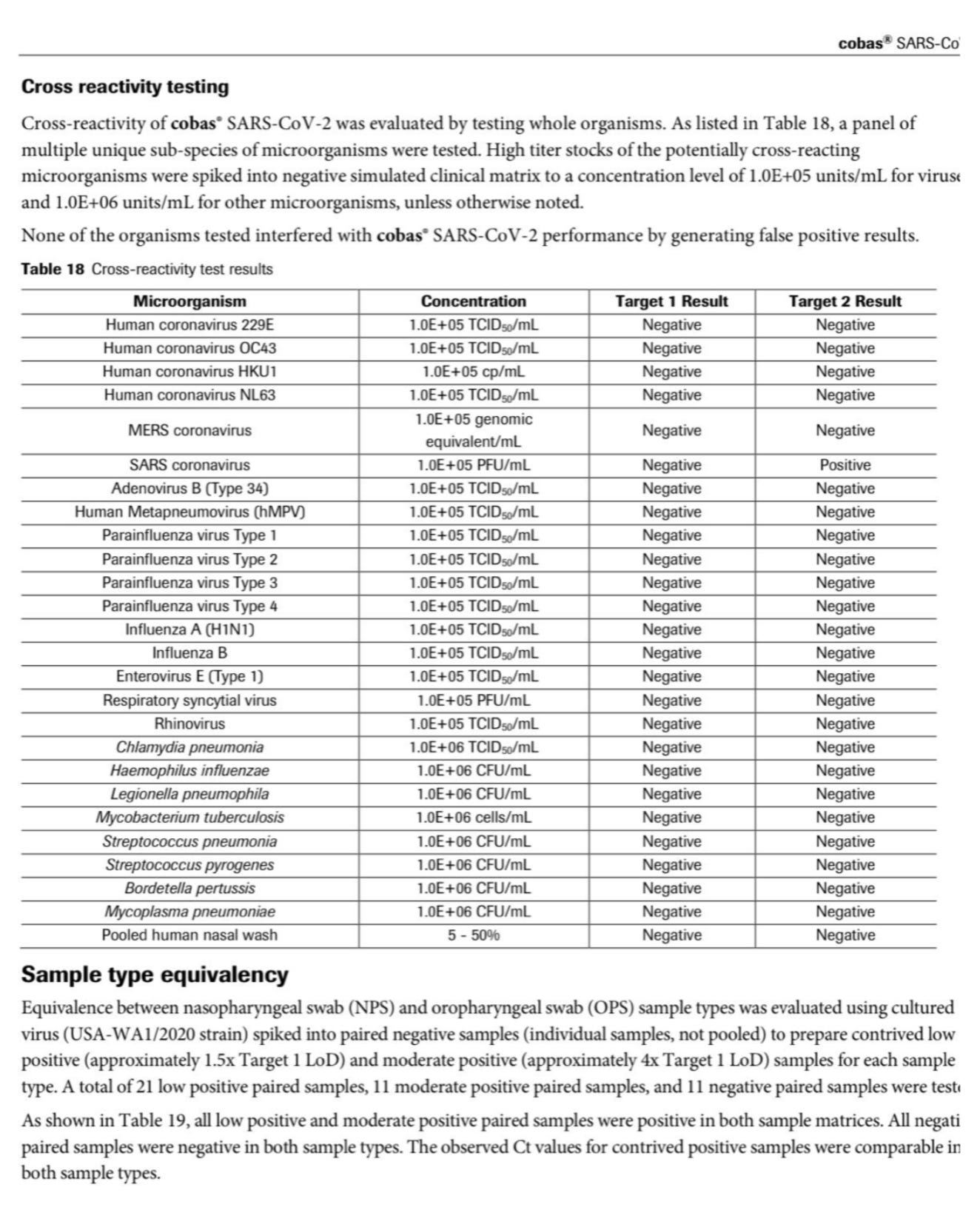
Here is the Roche Cobas Test.
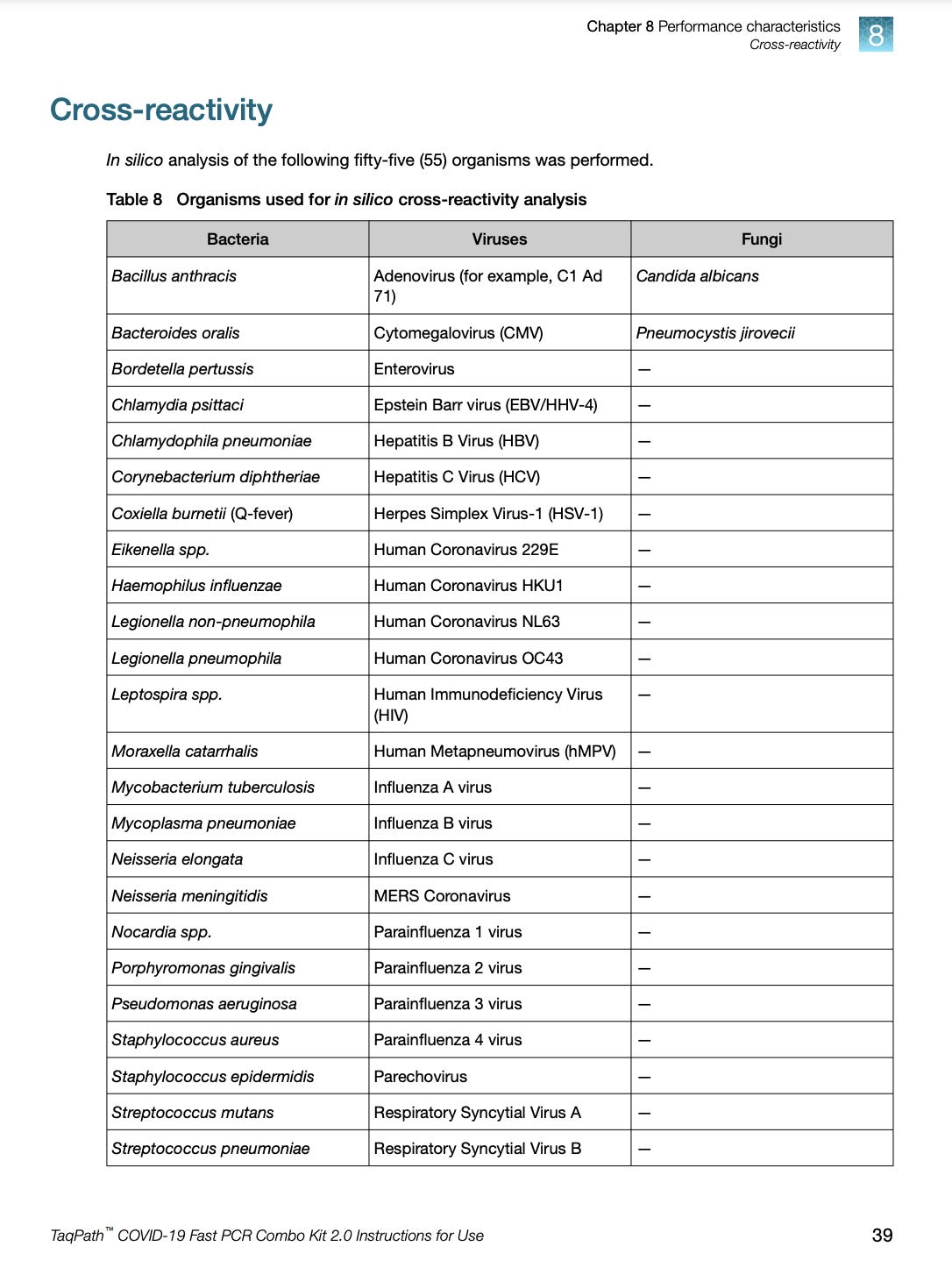
Here is another test that took the in silico approach.
You can see the handwaving about 80% homology and primer proximity on the next page.
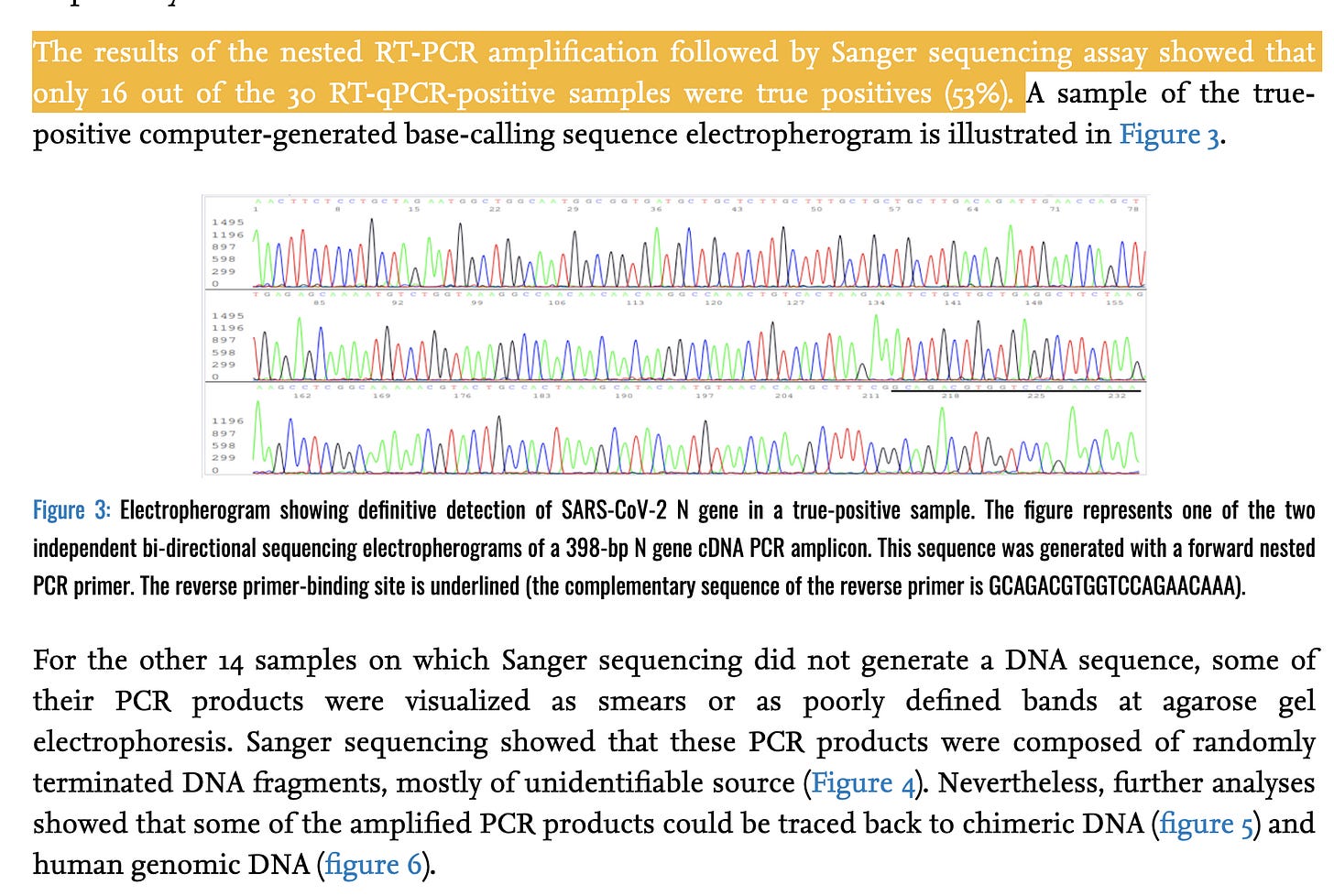
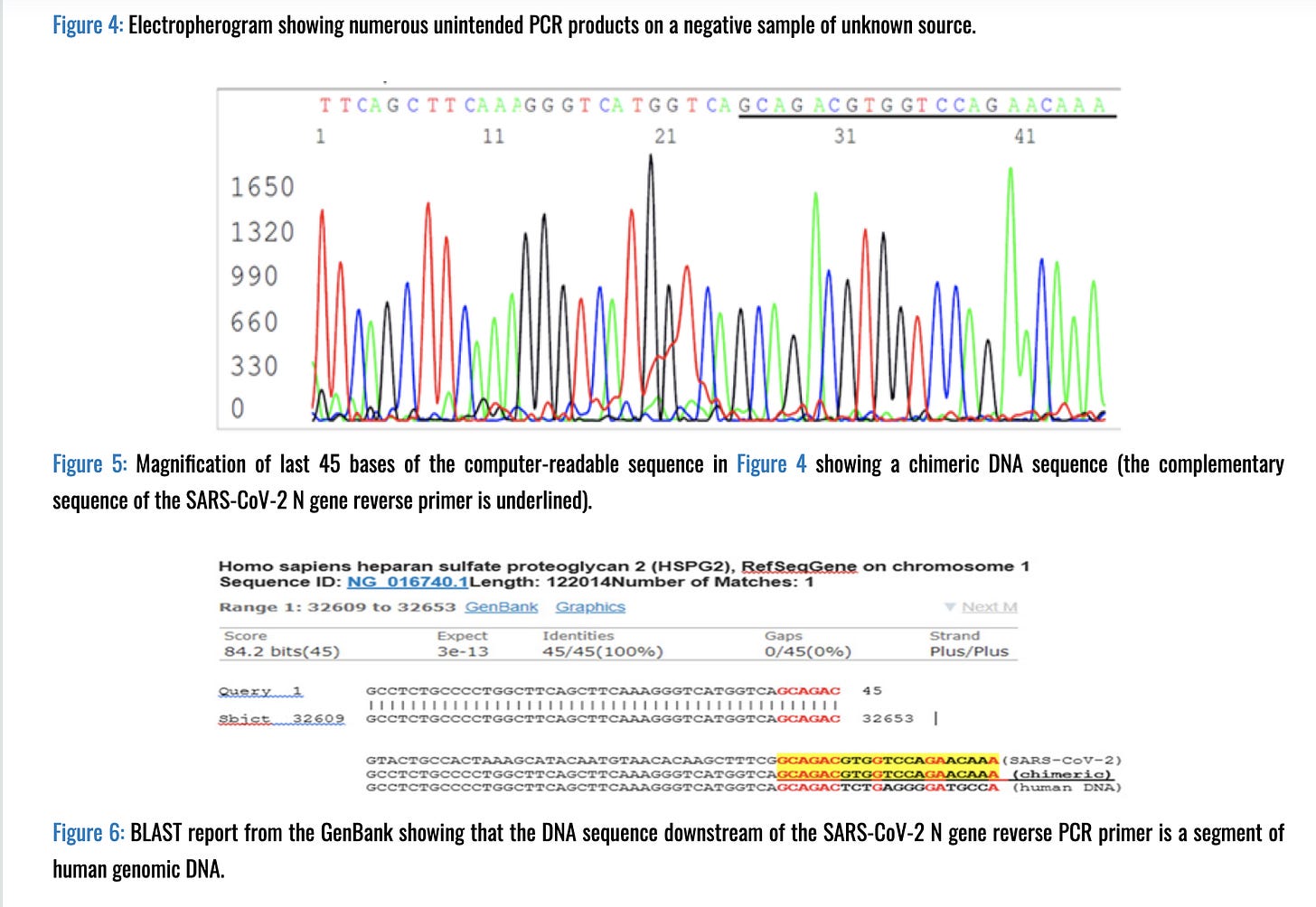
And to cap it off, here is a paper from Dr. Sin Lee that sequences the actual qPCR products to identify what they actually amplify.
And when sequencing is performed on these false positive signals, some Human DNA shows up.
It is unlikely this is happening in all test on the market and at low CTs but the likelihood of this happening at late CTs is real and very few labs will sequence validate anything after CT 32 for C19 testing. CDC recommends CT <= 28 for sequencing. This is important as the religious defenders of PCR perfection will always whip out the sequence validation card but they rarely ever have sequence validation on late CT samples used to quarantine people.
Its been 2-3 years of “Emergency use” of these billion dollar product lines. The cost to perform a live organism test would require ~50 organisms to be purchased, cultured and RNA/DNA prepped for qPCR at $300 per organism ($15-$20K).
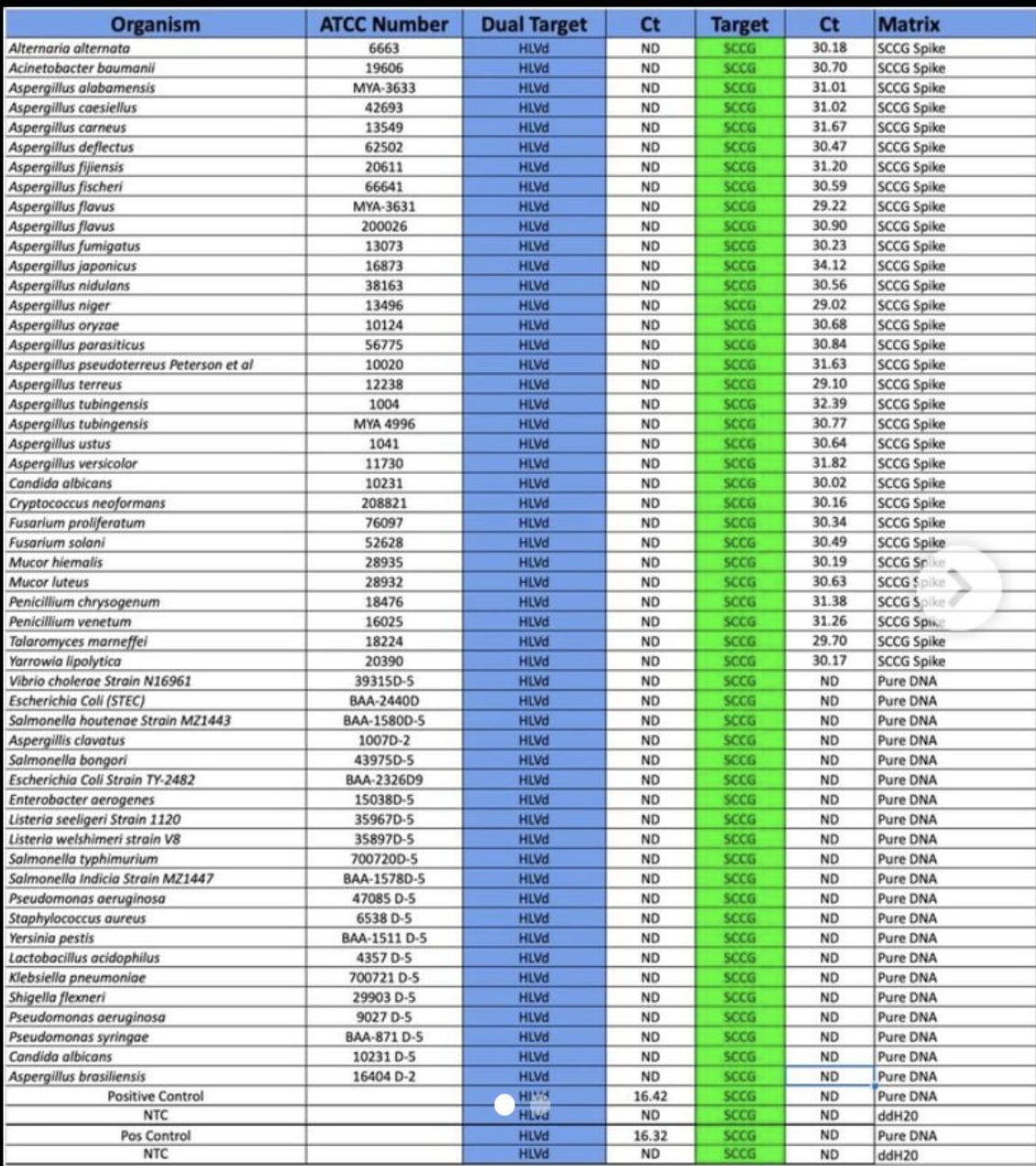
Even small companies operating with limited banking in the Cannabis space can pull this off for viroids that are no threat to humans but pose a significant risk to Cannabis cultivation. Below is our exclusion testing of an RT-qPCR assay for Hop Latent Viroid.
Look Mom.. No handwaving about 80% homologies.
The inclusion analysis (organisms you should hit) gets very tricky when you cant culture the organisms or no biobank has it yet. So novel pathogens like SARs-CoV-2 had to resort to synthesizing a DNA proxy to test the assays on. Many in the “Virus doesn’t exist” camp like to call this cheating as you didn’t ‘isolate’ the virus but this is a fallacious argument given synthesized DNA/RNA is the most purified form of the virus you can make. Even more pure than a protein coated mRNA and simple transfection of these nucleic acids have been proven to be replication competent.


RT-qPCR assays that target the world of RNA are best wet validated as the ‘in silico’ computer simulations don’t always capture the complexity of splicing and lariat formation in the transcriptome of all potential background organisms.
The moral of the story is that when “Emergencies” part the Red Sea of Regulations, companies begin to manufacture emergencies instead of manufacturing good products.









In PCR, primers don't really need to have unique genetic sequences, only PCR probes do.
Thanks for the information. Seems like the bigger problem in the cannabis industry is that the majority of testing labs and assay producers don't post full validations for their hop latent viroid assays at all. Would be nice to see complete metrics, not just cross-reactivity, for each assay so they can be more easily compared to each other.