CHD covered a recent paper from Dr. Brigitte Konig’s labs
It’s a good read with some important points regarding where in the manufacturing procedure these measurements are being made.
There are a few points that could use further testing and clarification that do not change the thrust of the paper but perhaps refine the magnitude of the contamination.
First point.
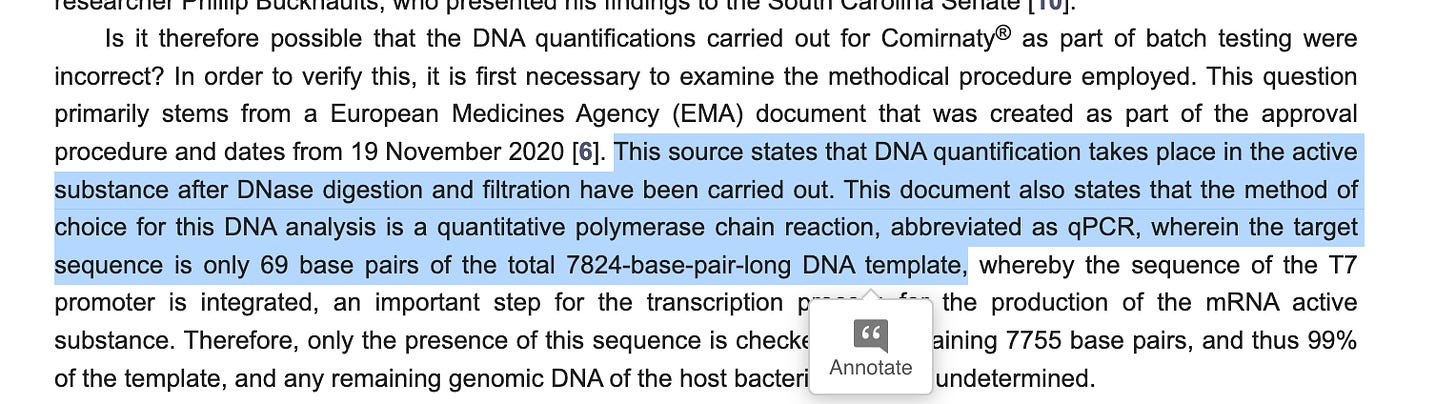
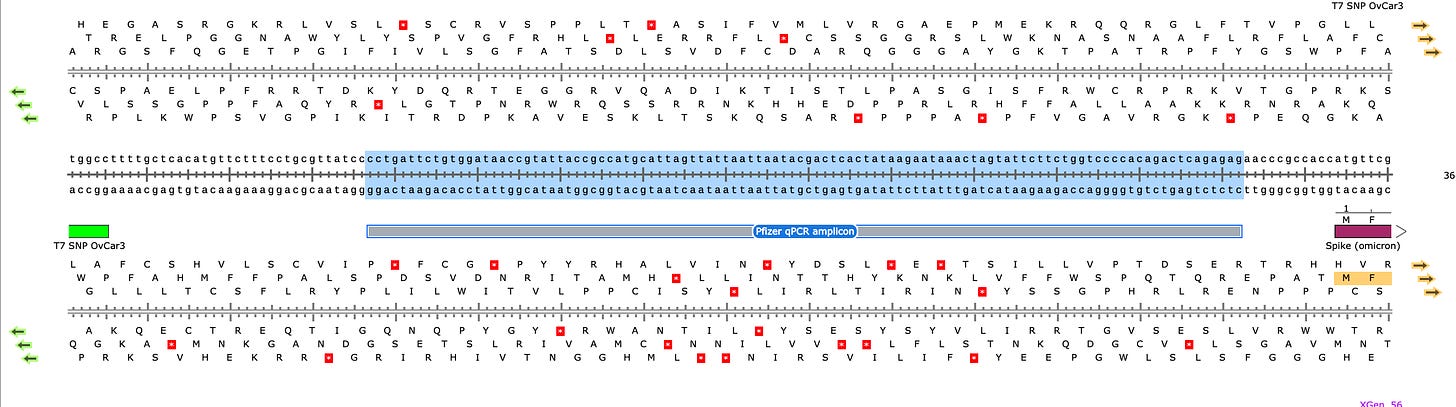
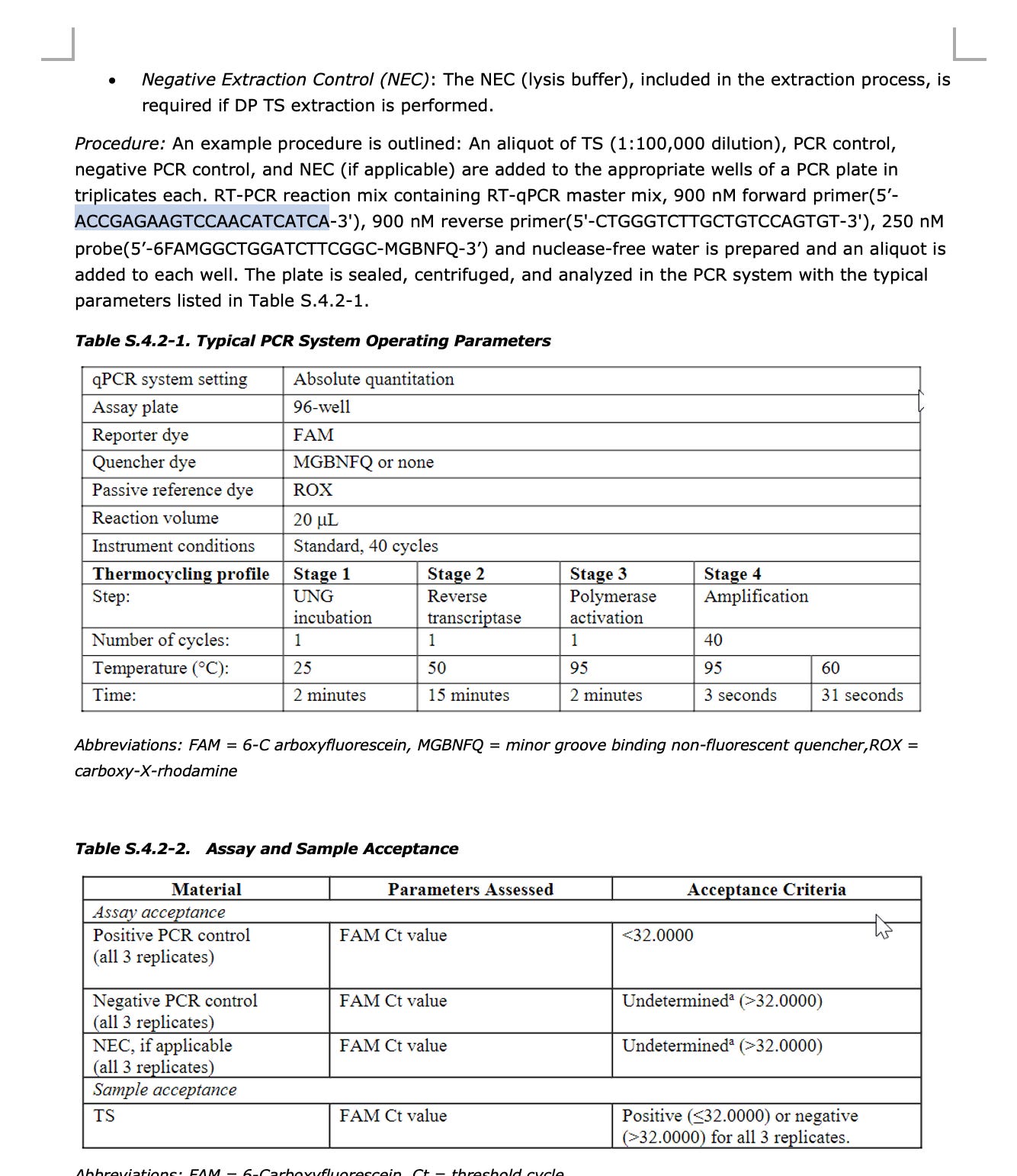
The documents we have on hand have Pfizer qPCR primers using SYBR green (no TaqMan Probe) and the amplicon is 106bp long. I pulled these from an EMA document that is covered on one of my substacks. I don’t know where they got the 69bp amplicon from but the longer the amplicon, the more it under-quantitates the DNA when DNaseI is involved. It does rest on the most highly transcribed part of the DNA which is a great way to suppress the signal.
Here is the EMA document describing these primers.
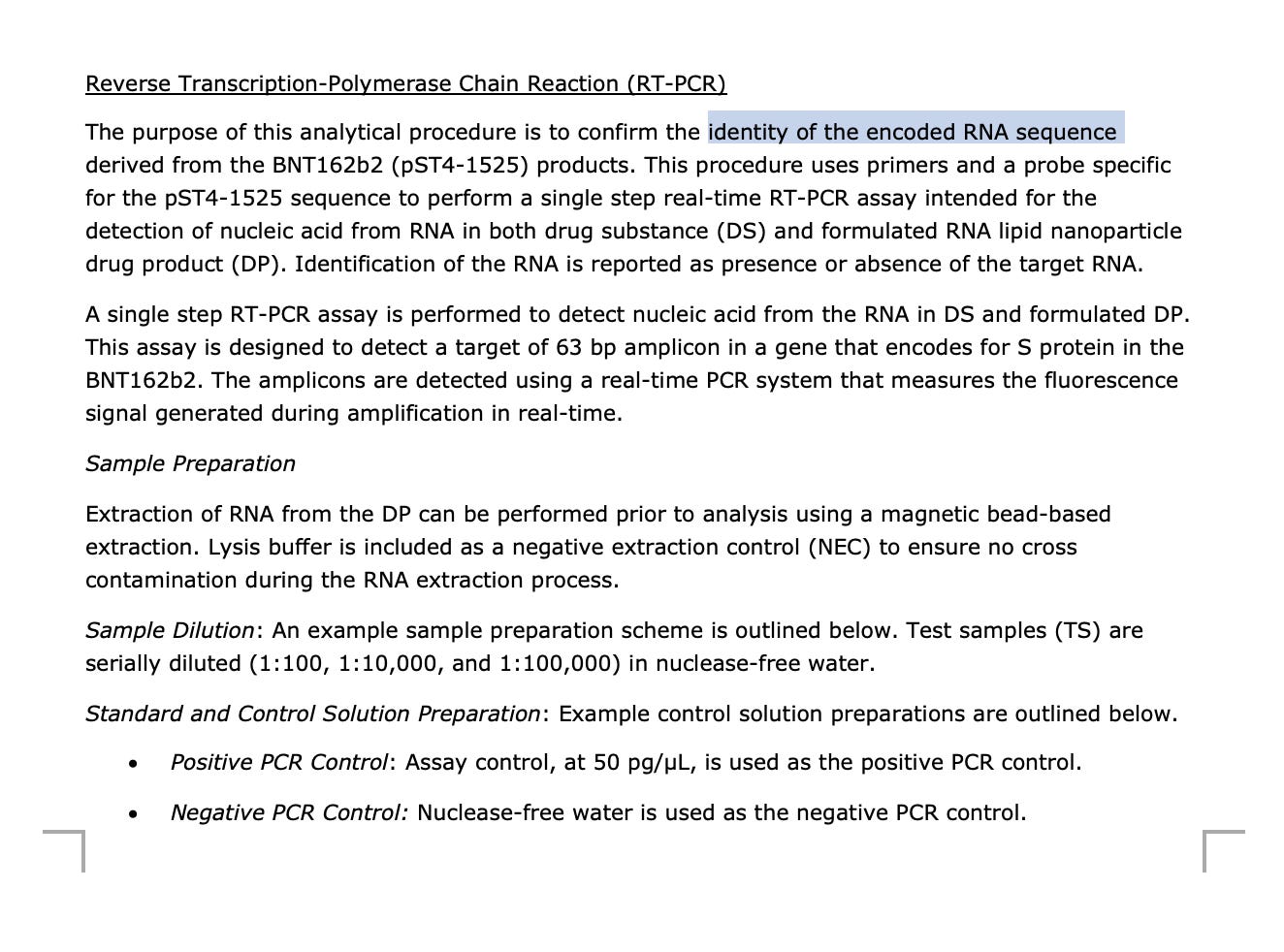
Note, there are 2 other primer sets described in the EMA documents. One Primer set lands on Spike and is used as an RT-qPCR assay to identify the mRNA. It is a quantitative TaqMan assay. This is a very important point as it shows they had a qPCR primer set that could quantitate RNA but chose to NOT use it and instead reflex to Fluorometry for RNA quantitation. This is NOT the primer set used to quantitate the DNA with qPCR. Those primers are shown above.
RT-qPCR Assay in the EMA documents.
There is a 3rd set of primers described in the EMA documents that pertain to ddqPCR to assess the polyA tail.
These are NOT the primers used to quantitate the residual DNA in the vaccines.
The only primers listed for qPCR target a 106bp region of DNA and as Konig rightly points out since this qPCR method only quantitates this 106bp region of DNA, it is assumed that the DNA for the rest of the ~7,718bp plasmid is in equal quantity. So any quantitation of this region needs to be mathematically adjusted for the fact that is only ~1/74th of the DNA in the vial (7824/106).
This math adjustment was likely true prior to DNaseI treatment. This is not true after DNaseI digestion as DNaseI digests the vector DNA more readily than the DNA that can hybridize to RNA in the Spike region. This is a known feature of DNaseI. It does not process RNA:DNA hybrids (I have another substack that covers this).
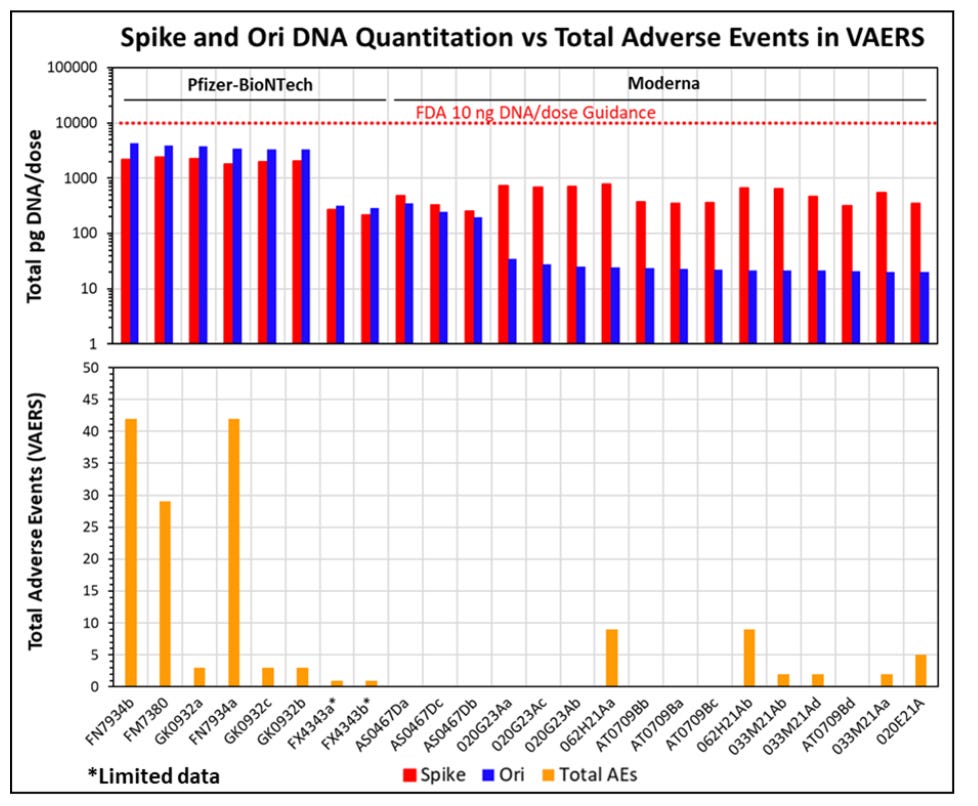
This was shown in Speicher et al where the qPCR signals for Spike were ~6CTs later than the Vector for the most (but not all) of the Moderna products. Its odd that all Moderna products do not do this. This suggests changes in their manufacturing and DNaseI steps and inconsistent manufacturing.
qPCR will also not pick up E.coli DNA or other non-plasmid DNA which does not have the target sequences used for qPCR. We did check a few lots with E.coli specific qPCR and found very late signals (>CT30) in some vials. We decided to ignore this low level as it cannot make an impact on DNA quantitation of the vaccine DNA at a CT of 20.
This is covered in Konig’s text and aligns with our results and Moderna’s own patents which state qPCR underestimates the DNA quantitation problem.
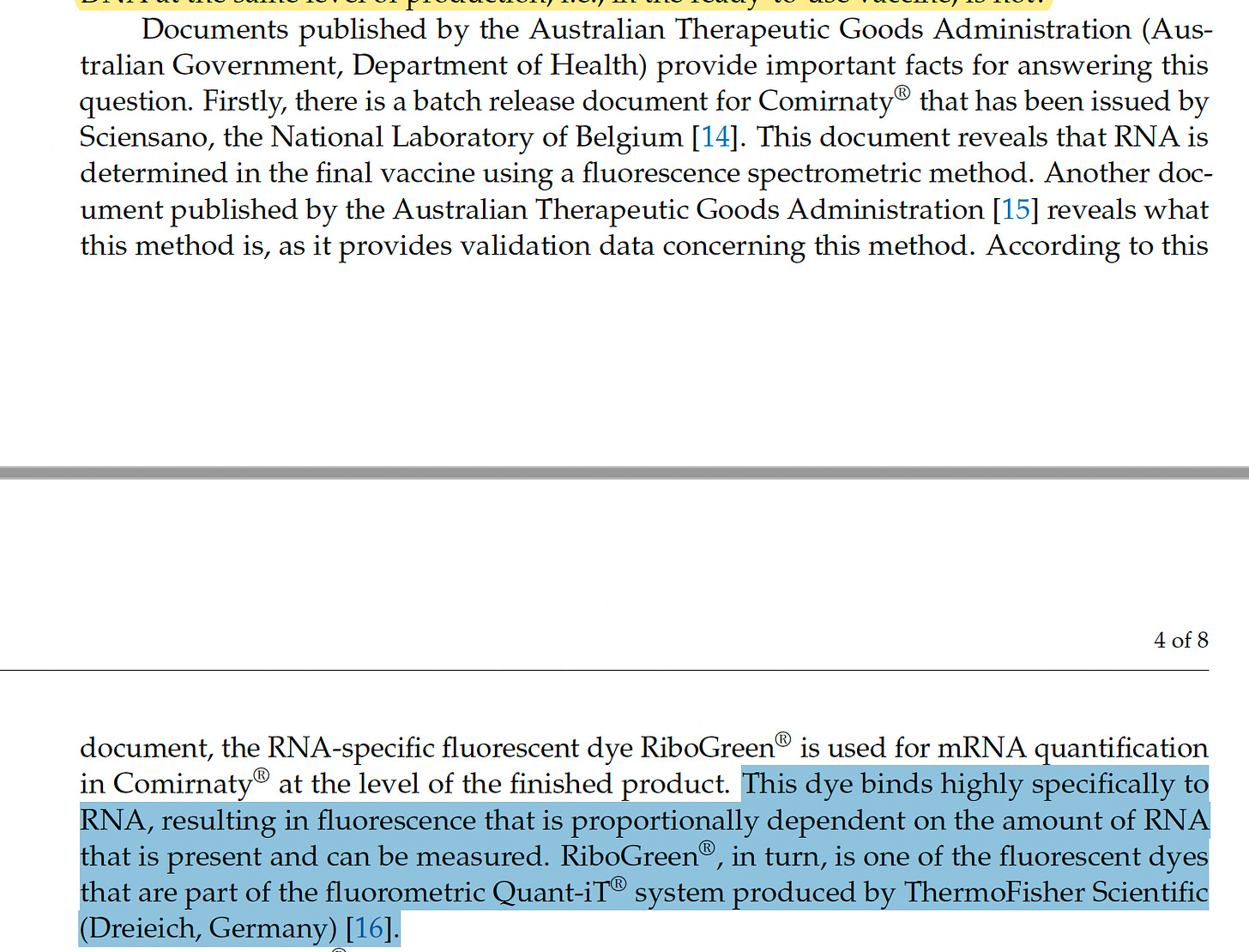
A very important point that doesn’t get any air time is that the measurements the regulatory bodies believe are being made by the manufacturers do not measure the final product but instead make a measurement at an upstream step.
This goes on to point out that if Pharma can measure the RNA at the final step, why not the DNA?
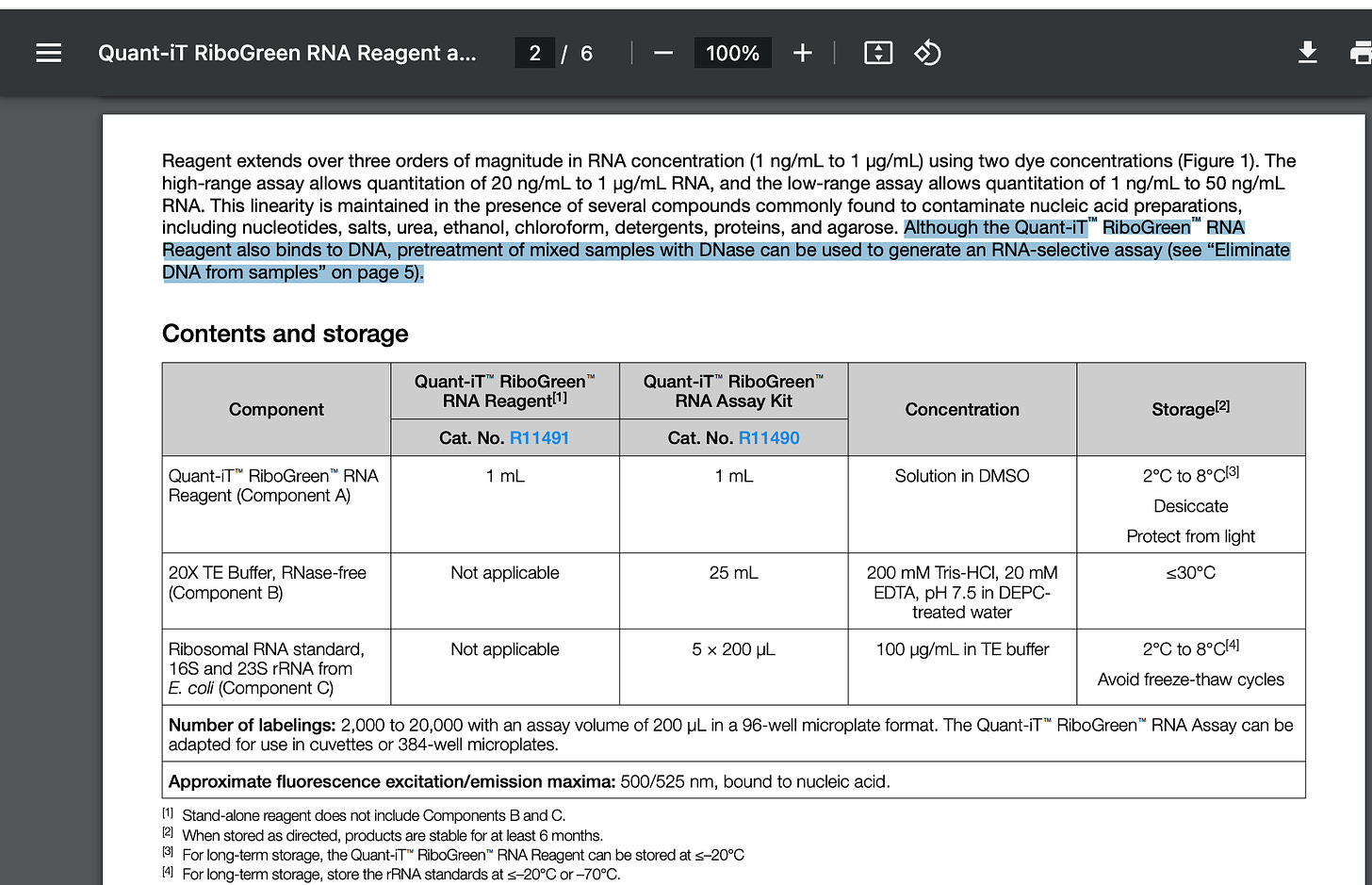
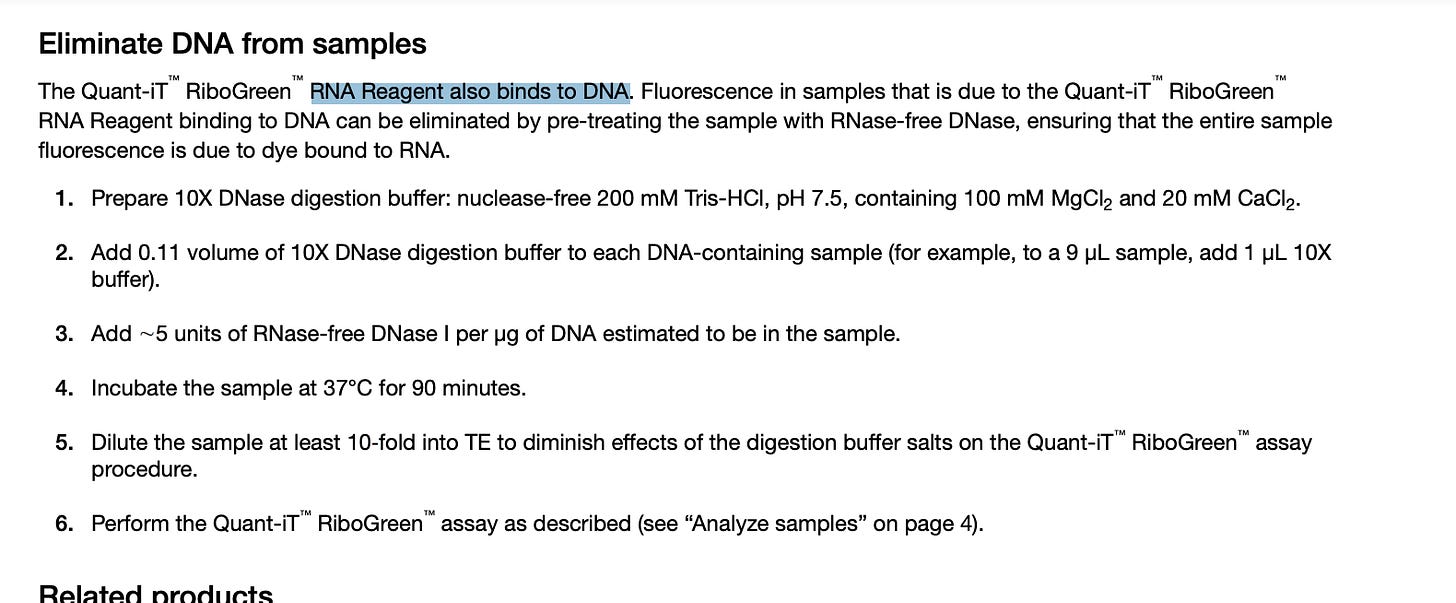
This is where one flaw in the paper emerges. RiboGreen is NOT a RNA specific dye. It in fact binds DNA 2X more than RNA and the protocol recommends the use of DNaseI before using it to assess RNA quantity. Why is it called RiboGreen? Because PicoGreen binds DNA over RNA 100:1 so RiboGreen is RNA favored at 2:1.
Jones et al covers this and is highlighted in our Deep Dive on Fluorometry.
The manufacturers protocol specifies the need to use DNase to generate RNA-selective assays.
Konig et al then go on to correctly point out that the RNA and DNA should both be measured at the same end point in manufacturing using the SAME tool. This is a point we have been raising for nearly a year now in two previous preprints.
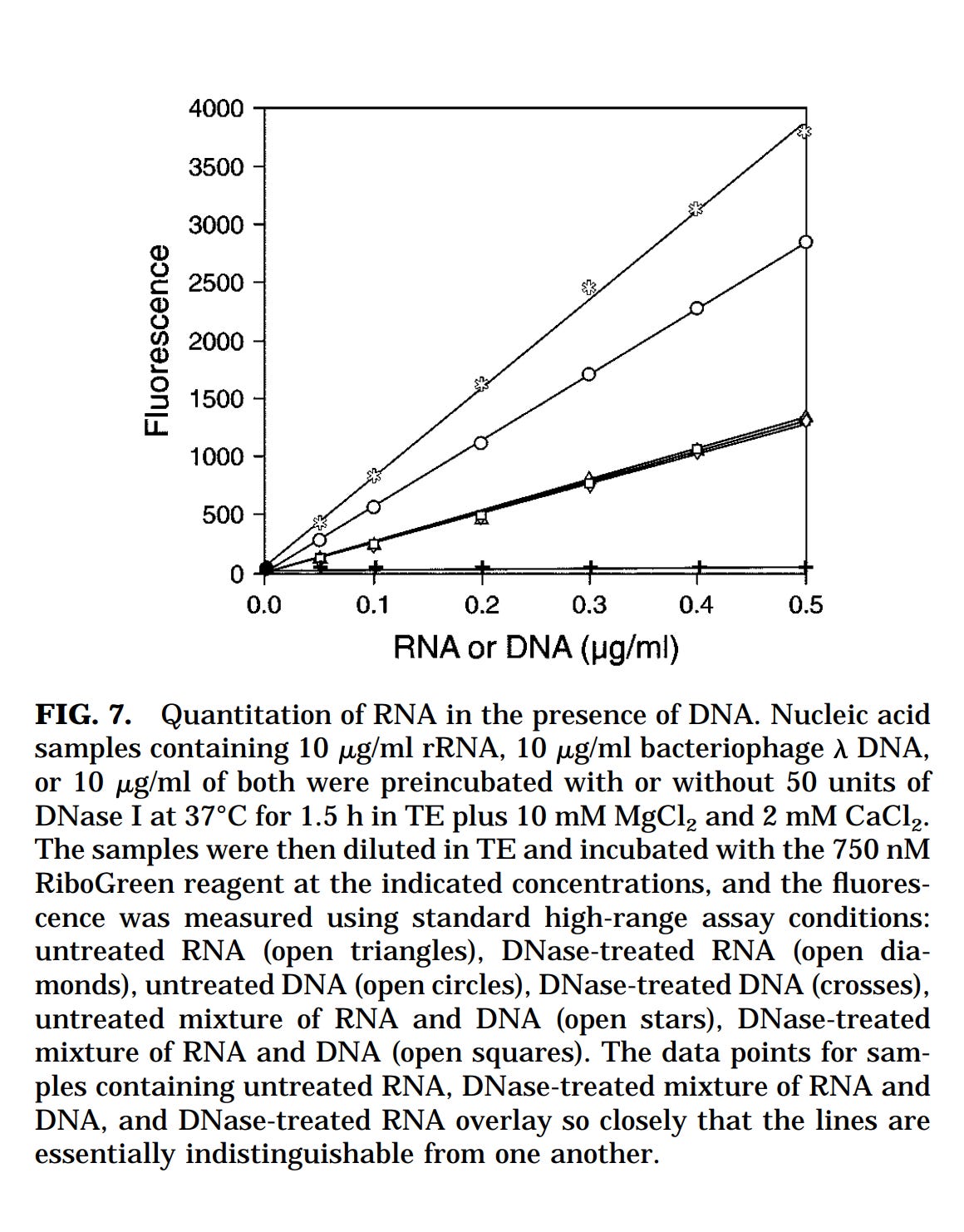
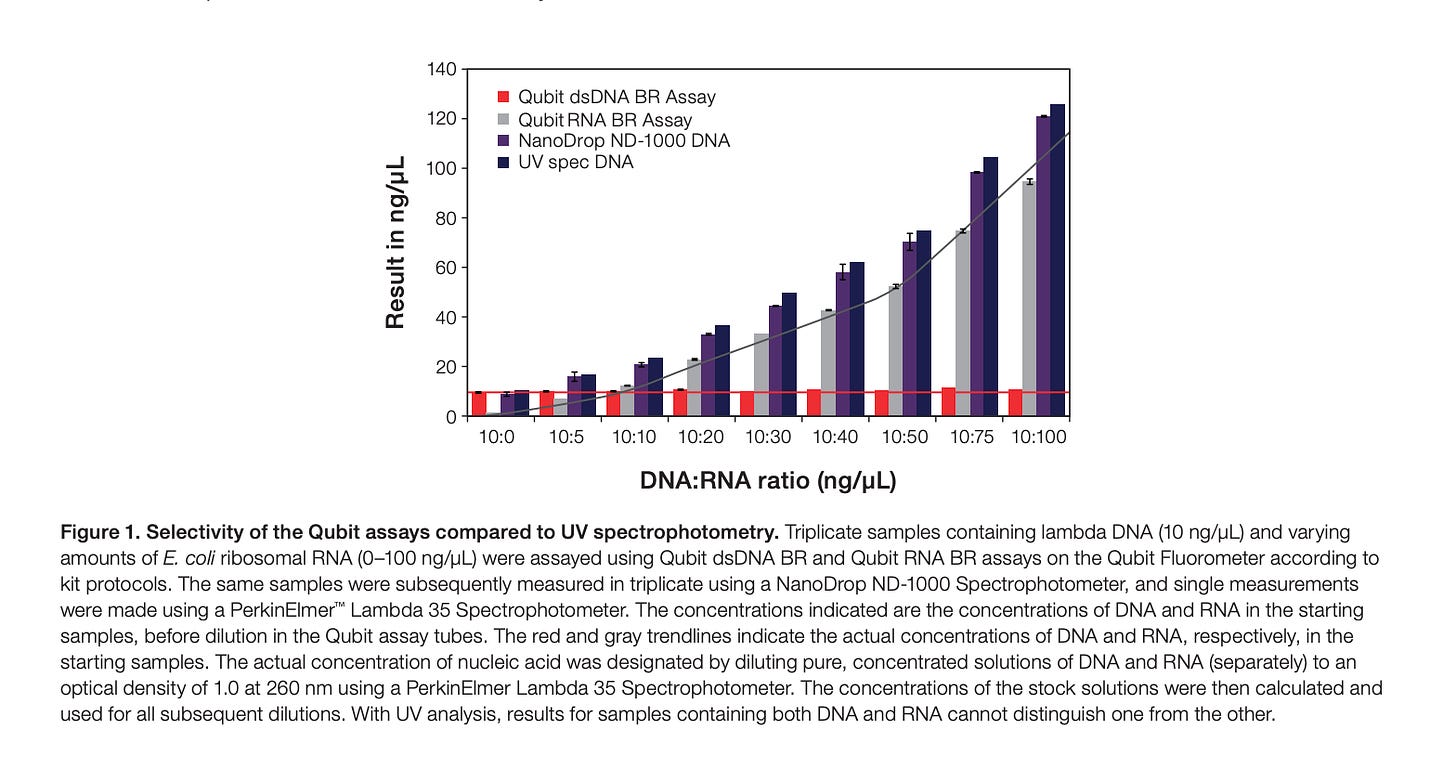
This is where the authors make one flaw. They resort to Thermo’s marketing material on this kit regarding PicoGreens specificity for DNA over RNA staining. I used to work at Life Tech in the genetic analysis department that made this technology so I know the reason the chart in this marketing material is not appropriate for this use case.
Below is the chart from Thermo (They acquired Life Tech). You can see the X axis only contains 10:100 DNA to RNA contamination and finds no influence of the contaminating RNA on the measurement of DNA with picogreen.
This is 1:10 contamination of RNA (10 fold more RNA than DNA) into DNA. What is required is a 1:3030 as that is the metric the EMA is asking for. They want less than 1 DNA molecule for every 3030 RNA molecules and I can assure you that when the RNA gets this high, it cross talks into your DNA signal.
They are simply resting on this manufacturers chart which is 2 orders of magnitude away from the contamination range we are looking at with these vaccines. They would need a chart that goes out to 10:30,300 on the X axis. They do not have this.
In addition, this chart from LifeTech was made using natural RNA. Any RNA that behaves more like DNA would nullify this ‘Validation’. That would include N1-methyl-pseudoU RNA and GC rich RNA (the vaccines have both) as this type of RNA hairpins more and thus forms more minor grooves for the PicoGreen dye to bind.
Second author Kirchner (Konig and Kirchner) voiced concern of our use of a generic PicoGreen known as AccuGreen claiming this manufacturer didn’t have the chart above. PicoGreen is off Patent so many other manufacturers can now make the same dye.
The best way to settle the concerns raised by Kirchner is to use an RNase (RNaseA from NEB) to eliminate the RNA so there is no potential for the dye to cross talk. This was performed in our Fluorometry deep dive substack and is in the process of being submitted for peer review. This prior was performed using AccuGreen. Our first preprint explored RNaseA with qPCR as the measurement. We are in the process of updating Speicher et al with RNaseA data with Fluorometry.
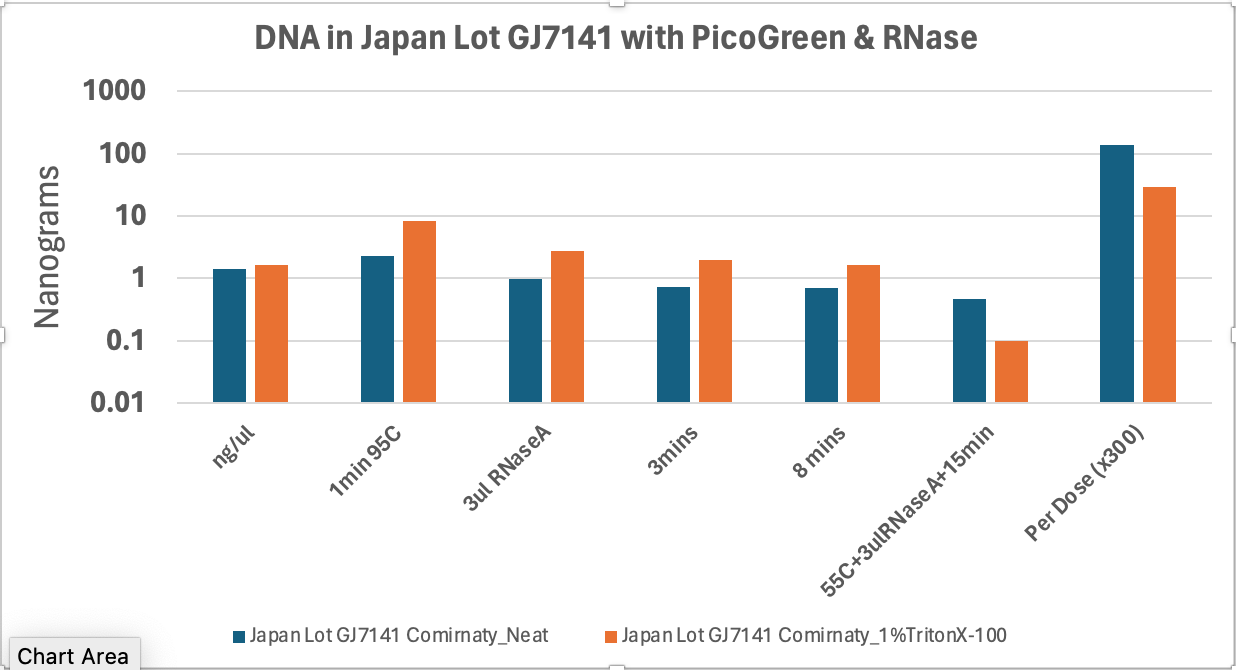
To evaluate this concern, I ordered a PicoGreen kit from Thermo and repeated this with our cleanest vaccine vials from Japan.
Just add 10ul of the vaccine directly into 190ul of the Qubit-PicoGreen reagent and you can see under 2ng/ul of ‘DNA’ (blue bars). Add in 9ul of vaccine and 1ul of 10% TritonX 100 to get to a final of 1% TritonX (Orange bars) and the numbers go up but this requires 95C heat for 1minute to really dissolve the LNPs.
A full dose is 300ul so 300X these numbers are seen on the right. These are cleanest lots we have (least DNA). It is interesting that the TritonX-100 signal degrades more with 55C and more RNaseA. This implies the PicoGreen can get into some LNPs but the RNaseA (much larger molecule) cannot until the detergent and heat are applied.
As you add in 3ul RNaseA from NEB, you can watch the ‘DNA’ disappear. This proves the Dyes are cross talking and what you think is DNA is really RNA elevating your signals. It is generally good practice to eliminate DNA or RNA when ever using these intercalating dyes as the dyes are not as specific as enzymes that have evolved to stringently differentiate RNA from DNA over millions of years. You can see the numbers come down ~10X once the RNA is removed.
As a result, I think the Konig estimates are a ~10X overestimation. Even if you discount this inflation, their lots will still be over the limit so their 500X estimate may really end up at 50X once RNaseA is utilized. So this critique doesn’t change the Alarm bells that need to be rung right now.
Those alarm bells should ring even louder knowing LNPs are in the picture and the FDA limits were derived from naked DNA contamination that has a 10 minute half life in the blood. Once LNP protected, this degradation cannot be relied upon and the limits need to be much lower.
This is also a good place to remind people that the DNA doesn’t need to get into the nucleus to create havoc.
One other caveat. We have seen lots vary 1,000 fold with qPCR using the same qPCR assay and the lots we have from Germany are the hottest lots we have seen yet (CT 13). This is why I am using ‘~10X’ estimates as no one to date has measured the same vaccine lots with different methods. We are all working with what we have on hand without much lab to lab overlap in lots or vials.
We have also seen qPCR estimates vary 1,000 fold based on using primers that land in Spike versus primers that land outside of spike (See Moderna’s lots with Speicher et al). So Pharma has all the levers they need in order to pull the wool over the regulators eyes as long as they are allowed to measure the RNA with meters (fluorometry) and the DNA with inches (qPCR).
There is no excuse for the valid critiques Konig et al raises regarding testing only the RNA as the final product and the DNA at some irrelevant step earlier in the process. They should both be measured with Fluorometry + RNaseA (for the DNA) and Flourometry + DNaseI (for the RNA) to assess total content of DNA contamination independent of its size.
The 200bp size restrictions they have in the regulations are irrelevant once LNPs are in place. This was a size they deemed easy to degrade once injected but this DNA is getting directly transfected without degradation so they can no longer rest on the small sizes being easily degraded.
Use of a single qPCR assay was never acceptable with COVID testing but it somehow is acceptable with the mandatory liability free injections they are forcing into children and pregnant women.
The ATIP (FOIA) emails that
revealed, show the regulators have never measured this themselves and are relying on the honesty of the Pharma firms that just demonstrated they will deceive them with SV40 sequences (Pfizer only). This can only be described as an immense Cover Your Ass move and it appears they may be too big of an Ass to cover.







also this: https://www.theepochtimes.com/world/health-canada-withholds-whether-it-asked-pfizer-to-remove-sv40-sequence-in-covid-shots-5652007
Great article, will no doubt be ignored by the lying, criminal, mass-murdering gov "scientists", "MDs", and "administrators" that get paid $billions of our tax-dollars to murder us, while they ignore reality almost 100% of the time and pay the press to repeat their lies.