In order to more efficiently search for genome integration events, we have designed a target enrichment system that can fish out DNA needles from a haystack.
Target enrichment is something we do routinely at Medicinal Genomics. It is a method that allows you to focus your sequencer on desired regions of the genome you’d like to sequence.
Exome sequencing is performed using these methods. 1% of the human genome is coding for proteins and exome sequencing focuses the sequencer on these targets of interest.
In cannabis breeding, people are moving more toward whole genome sequencing as the sequencing costs come down but if you want to find ‘needles in a haystack’ (rare events that are not present in every cell), WGS is expensive.
Most sequencing performed today is performed on collections of cells that all share the same genome. When you want find a rare variant that only exists in a subpopulation of those cells, you need to sequence the collection of DNA to much higher redundancy to ensure the sampling of the rare allele. Single cell genomics has recently emerged to help address this field.
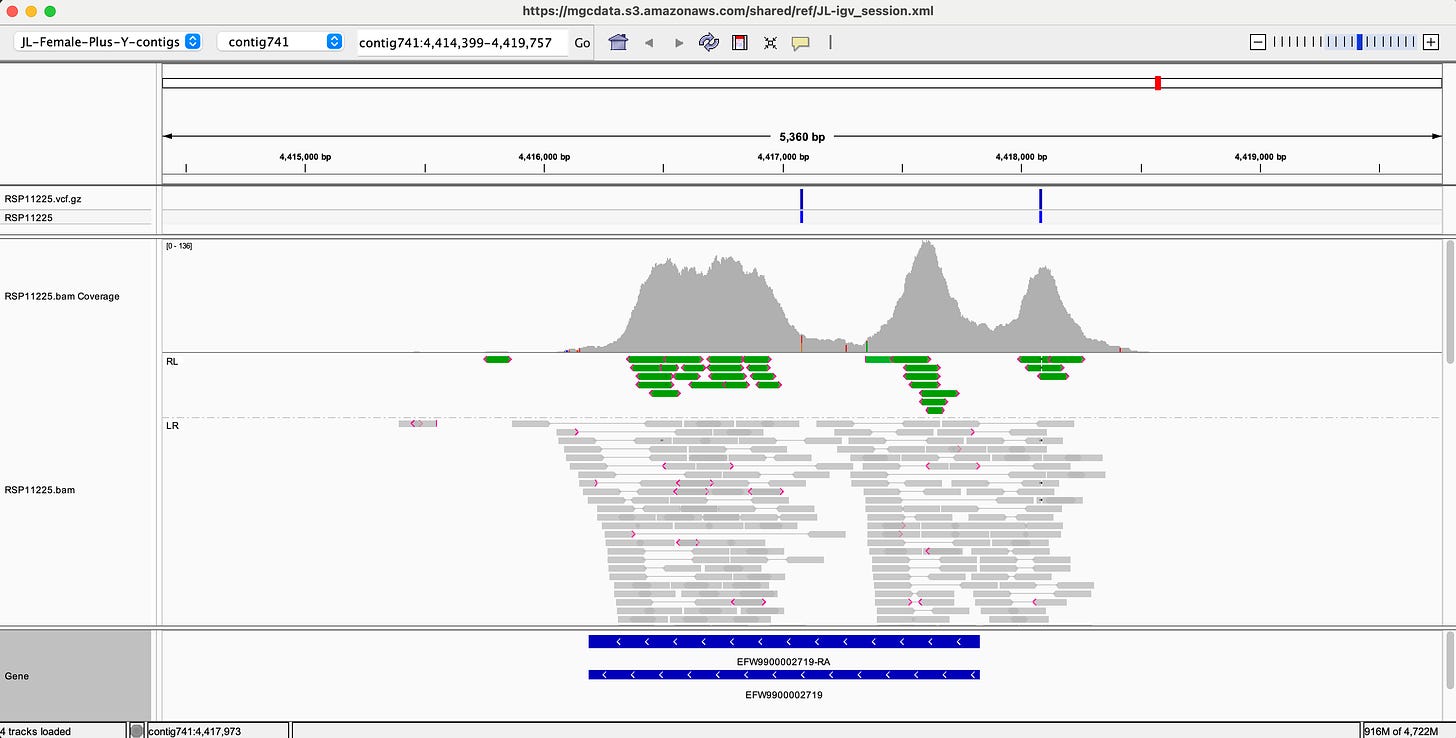
Here is an example using targeted enrichment on the Cannabis gene that synthesizes THCA (THCA synthase os THCAS).
Above is an IGV track. The sequencing reads are grey horizontal lines in the middle track. The depth of sequencing coverage is depicted as a histogram on the top track that looks like a mountain range. Its actual depth is depicted in the upper left corner of the track [0-137]. That means some bases are covered by 137 illumina reads. The blue lines on the bottom track is the annotated 1.6kb gene known as THCAS.
Compare this to the same gene sequenced with Whole Genome Sequencing.
You can see that the sequencing isn’t targeted over a predefined target of interest and the average depth of coverage is lower [0-66]. This process is targeting ~3.2Mb of the coding content of the cannabis genome. It is cheaper than WGS and often delivers higher sequencing coverage over your targets of interest.
So how do you Target? PCR is one approach but doesn’t scale well for million base targets. It can also become problematic in very polymorphic genomes like Cannabis. There can be a SNP every 25-50 bases in Cannabis and this can cause allelic drop out in PCR as you can’t anticipate all of these variants in your primer design. You also need to know both ends of a target molecule for PCR to operate and sometimes you can’t predict what one end of the molecule is going to be in advance when you are looking for structural variations or genome integration events.
To find an integration event, one end of your molecule might be spike while the other is chromosome 12. Since you cant know the location of the integration event in advance, you cant design PCR products that go hunting for this. You need a 1 primer system that can fish these integrants out.
Another approach is to use longer primers to hybridize out molecules similar but not identical to your target of interest. This approach is more capable of finding variants and integration events.
If you want a background on the general methods people use there is a description with ChatGPT that is on point.
What is Target Enrichment? By ChatGPT
For those who prefer visual references below is a figure that depicts this workflow. In this case we are targeting the plasmids that are in the vaccine. Since the Moderna and Pfizer vaccines only contain 13kb of sequence, we can target this with just 218 biotinylated probes. It starts with making a Whole Genome Shotgun library which shears up DNA (usually with DNaseI) and ligates Illumina adaptor sequences (Orange) to every DNA molecule. This library can then be hybridized against biotinylated probe sequences you want to pull out through hybridization. This step requires denaturing the library into single stranded form.
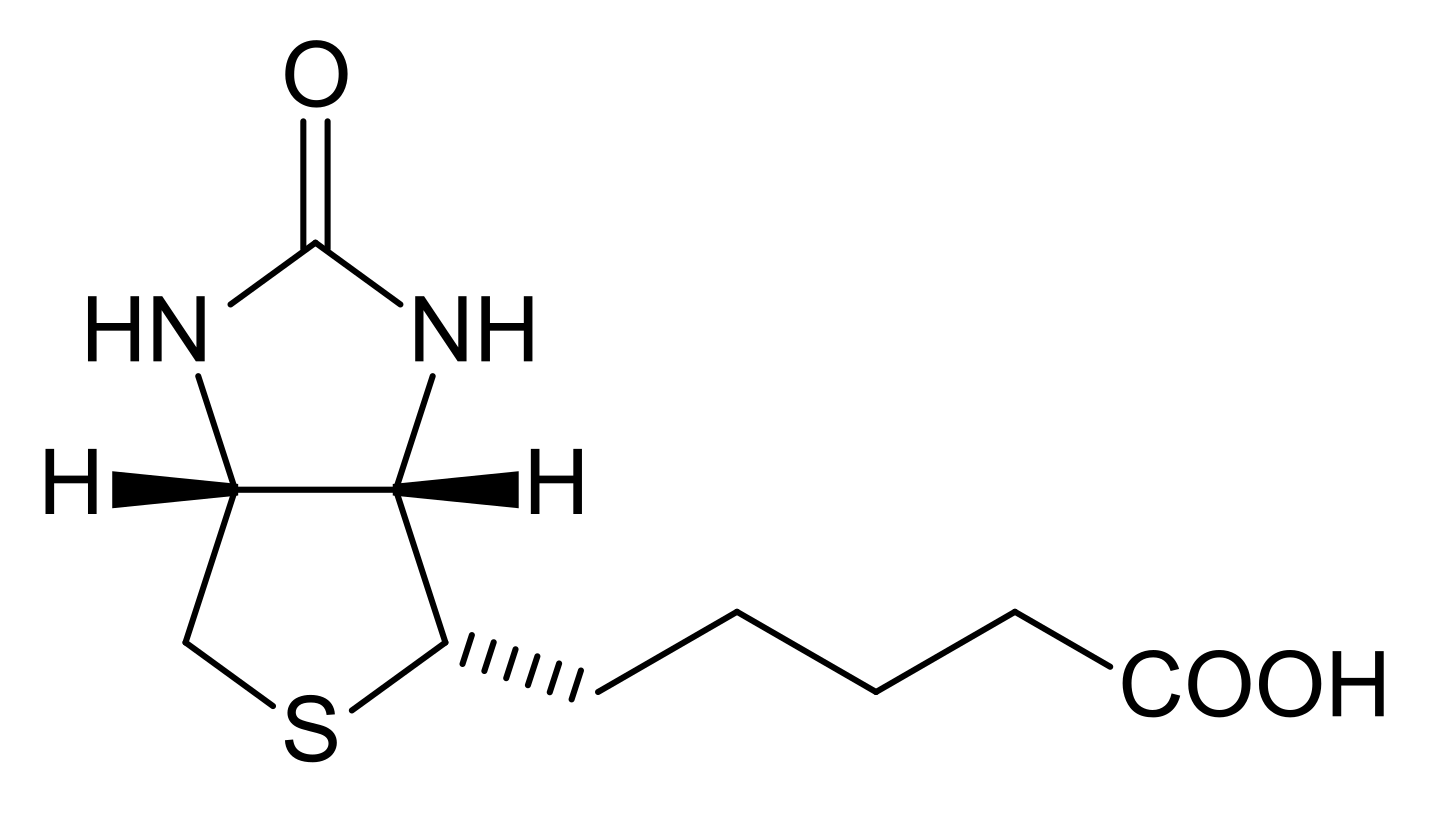
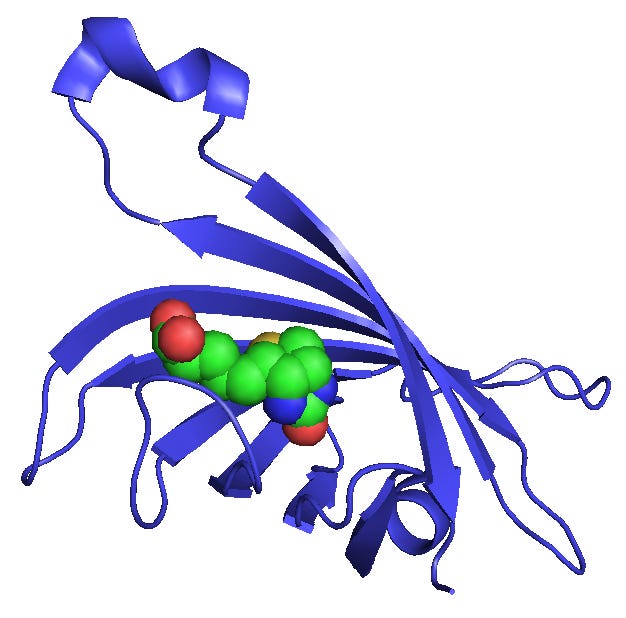
Biotin is an important genomics tool. This small molecule is easy to synthesize into an oligonucleotide or probe sequence. It serves as a molecular fish hook as Streptavidin strongly binds to this molecule under aqueous conditions.
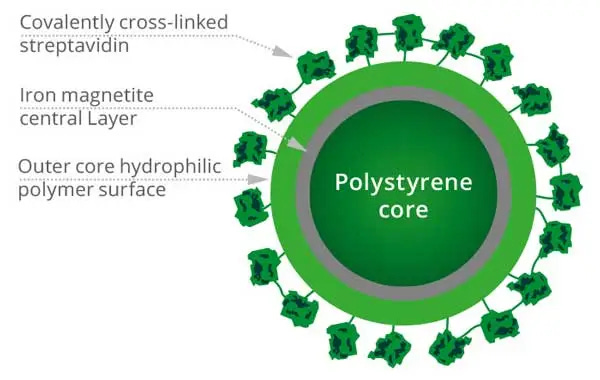
Streptavidin is a protein that is easy to couple to magnetic beads
This makes it very easy to hybridize your targets to biotinylated probes and fish them out with ‘Step Beads’ and a magnet.
Once you have collected these beads and have stringently washed them at 65C, you now have a collection of DNA in the library that have sequence homology to your fish hooks (biotinylated baits).
You can then put these beads into a PCR reaction and amplify the targets with homology to your probes or ‘baits’. As mentioned earlier, for this amplification to work, you first need to make a Whole Genome Shotgun library that has Illumina primers adapted to each end of the DNA as seen in the pictogram above. These Illumina primers can be used to amplify the DNA that the probes capture after you have rinsed the beads of DNA that doesn’t hybridize to your probe sequences.
You will notice that the methods include ‘Blocker oligos’. These are used in massive excess to hybridize to the Illumina adaptors. Since these sequences are shared between all library molecules and complementary, if you don’t block the Illumina adaptors, the library molecules will chain up recruit off target DNA. Some are often tempted to put these Illumina adaptors on after the baits have pulled out the DNA. Its possible to do this but you don’t alway know how much DNA (it will be low if you targeting a small region) you will recover and that makes the adaptorization process very tricky. There is also a preference to put the adaptors on first as you include DNA barcodes in the adaptors which allows you to put 10 patients worth of DNA into one capture process and sort out the multiplexing (de-multiplexing) via sequencing these barcodes. This brings the enrichment costs down 10 fold.
In the case of targeting larger regions of interest like 3.2Mb in Cannabis we use a target enrichment approach known as Agilent SureSelect. This utilizes a massively parallel photolithography system that can synthesize 100,000- 1 million oligos in parallel with UV light as the coupling ‘reagent’ during DNA synthesis. This has also been achieved with Ink Jet synthesis. One of the geniuses (Alan Blanchard) behind the SOLiD sequencer first described Ink Jet synthesis with Lee Hood.
Once the oligos are synthesized on glass, they can be cleaved, pooled and used for enrichment. They are usually limited to 50 bases in length with these massively parallel oligosynthesis methods. When you want to target smaller regions, we have been using IDTs XGen Lockdown probes which are 120 bases long and synthesized with more traditional ‘Caruthers’ DNA synthesis methods. These panels are more economical when the target space is small.
This may come in handy for breeders who are interested in just 1,000 SNPs in the cannabis genome and are very price sensitive. For instance, if you just want to confirm a plant is a clone of other, a cheaper 1,000 SNP panel will suffice and can be a ~1/3rd of the cost of the 3.2Mb panel. This can also be used for breeding projects that want to survey 1,000s of plants but don’t need 300,000 SNP resolution that the 3.2Mb panel offers.
If you need to find a needle in a haystack, targeting is the most read efficient means to do this. This is pertinent for rare variants such as plasmid integration events. These events will not occur in the same place in both chromosomes and they may only occur in a small percentage of the cells. This is a mosaic (only in some of the cells) haploid (only in one of the chromosomes) event.
There is a hilarious tweet from Debunk the Funk, claiming these could be easily found with a Southern Blot and therefor genome integration events would have been easily found by now.
This is sadly not true and displays his shallow understanding of this field. Southern blots are not single molecule sensitive. They can stain a large collection of DNA but they are terrible for finding a ‘needle in a haystack’ when the needle is in an unknown location in a minority of cells.
Whole Genome Sequencing would likely require 30-3,000X sequence coverage just to find events that hit 1-10% of the cells. This can require billions of Illumina reads to find. Targeting may drop the requirements 100 fold and enable much deeper sequencing of the regions of interest. So while you may pay $50-$100 to perform the capture experiment, you will more than make up for those costs in sequencing savings (thousands of $).
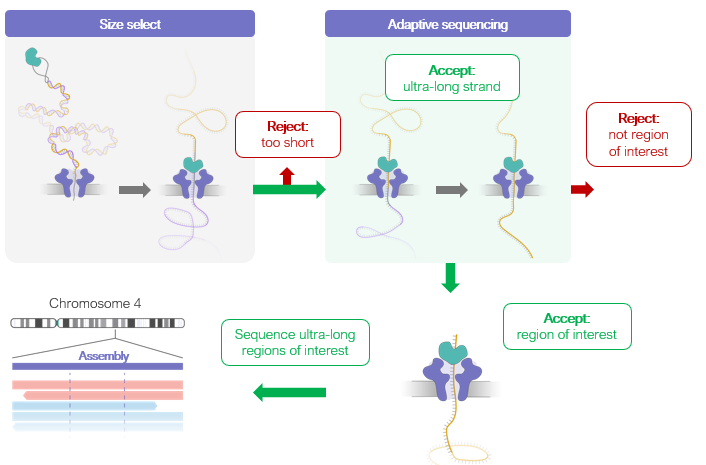
Oxford Nanopore Technologies (ONT) has come up with a clever means to enable this targeting directly on their sequencer chip. This is known as adaptive sequencing. Since Oxford Nanopore sequencers are real time sequencers, they can base call the molecules zipping through the pores at 250bp/sec and make a real time call to reject the molecule from the pore with a current reversal.
There is a sacrifice in that the smaller your target gets in the pool of DNA, the more time the sequencer spends partially sequencing rejected molecules. These sequencers also require 500ng of DNA (~78,000 cells at 6.4pg/ diploid genome) where as the Illumina sequencers can sequence single cells. The ONT sequencing is marvelous in that it has no read length limit but these features are currently more expensive than Illumina sequencing. The per base accuracy is also lower than Illumina but the ONT platform is quickly catching up with its 2D sequencing chemistry which sequences both strands of the DNA in order to error correct it.
Stay tuned. We are just testing out these targeting enrichment tools and if they are successful, they may offer a much cheaper means to screen tumor samples for vaccine induced integration events.
The probes required to do this are public on the link at the top of this thread.










Kevin McKernan is mission oriented, and introducing others to the targeted sequencing tools needed to detect low frequency genetic integration events. This is how we will find out what the contaminated mRNA jabs are doing in the bodies of the recipients. (Kevin is what researchers ought to be.)
Sharing is caring.
Thanks again for sharing you hard work.