There is a funny game of technological leap frog occurring in the genomics space.
I can recall the company meetings at Life Tech that heralded the coming of handheld biology in 2009.
This was Life Tech getting a wee bit excited that one of its $5,000 product lines (Qubit) was becoming a lab staple everywhere in the world. It was because it was small and cute and most lab equipment prior to this required a computer tied to a large plate reader. This big box model was getting in the way of sales as many universities had additional CapEx approval requirements for instruments over $10K. Any device you could make under this price point was a much more liquid sale and could be sold with credit cards online.
The Qubit above ‘democratized’ DNA quant. The word ‘democratized’ was the new business lingo. Now the word appears to be an insult to ones intelligence as democracies around the world have become democidal-crazy. But you get the point. Smaller foot print. Main-frame to iPhone mentality was hitting life sciences.
One of the products of this era was the Nanodrop. Everyone had one and everyone loved it and hated it at the same time. It was really a Quantum fit or QuFit.
Why the Love-Hate? Traditional UV specs required large volumes, quartz cuvettes and were frozen in time. Since the application hadn’t changed, the devices that ran UV spec had software frozen in Cobalt time. The Nanodrop turned the 500ul Cuvette into a 1ul observation volume. It was cheap, small and had no consumable reagents. The read times were seconds. As a small footprint UV Spec, it rapidly replaced all the dinosaur boxes.
So why the hate? While the Nanodrop mastered the form and fit for the market, the core technology was limited in the information UV spec could provide.
RNA and DNA have similar absorbance at 260nm so the platform can’t quantitate DNA vs RNA. It can only guess at total nucleic acids and anything else that absorbs in the 260nm spectrum will inflate the readings.
For pure oligos that come off of a DNA synthesizer, it’s perfect. For complex biological samples, it’s a ball park at best. Some labs use the ratio of 260nm vs 280nm absorbance as a measure of the protein contamination in a DNA/RNA extraction as proteins tend to absorb in the 280nm range. 230nm is often used to assess phenol or other contaminants that might come through the prep.
The take home message is that this read out is NOT specific to DNA or RNA. It’s a UV absorbance of all molecules present and I have no idea what LNPs, ALC-00315 or other ingredients in the vaccine may do to these readings.
So when I saw the EMA documents speaking to UV Spec as a final read out for what is injected into people, I cried.
The instrument that all genomics jocks least trust was now the arbiter of liability free, mandated injections.
It’s like if you were about to go under for open heart surgery and the doctor shows up with a toilet plunger.
We posted on UV Spec data back on March 15th. This post looked at vaccines that were purified with a CTAB/SPRI isolation procedure and as a result we lost a lot of the small fragments in the DNA purification process but we eliminated the LNPs.
Yes… I have spent much of genomics career perfecting size exclusion magnetic bead purification technology. If you want to selectively remove small fragments of DNA, there are many ways to do this with SPRI. Moderna even mentions (in their patents) the use of some of the techniques our team at Agencourt perfected but it’s not clear to me Pfizer knows how to use them. Moderna is generally cleaner from a DNA contamination standpoint.
David Wiseman asked a question. What happens if you use UV Spec to measure the final Drug Product in the LNPs as Pfizer claims to be doing for the EMA?
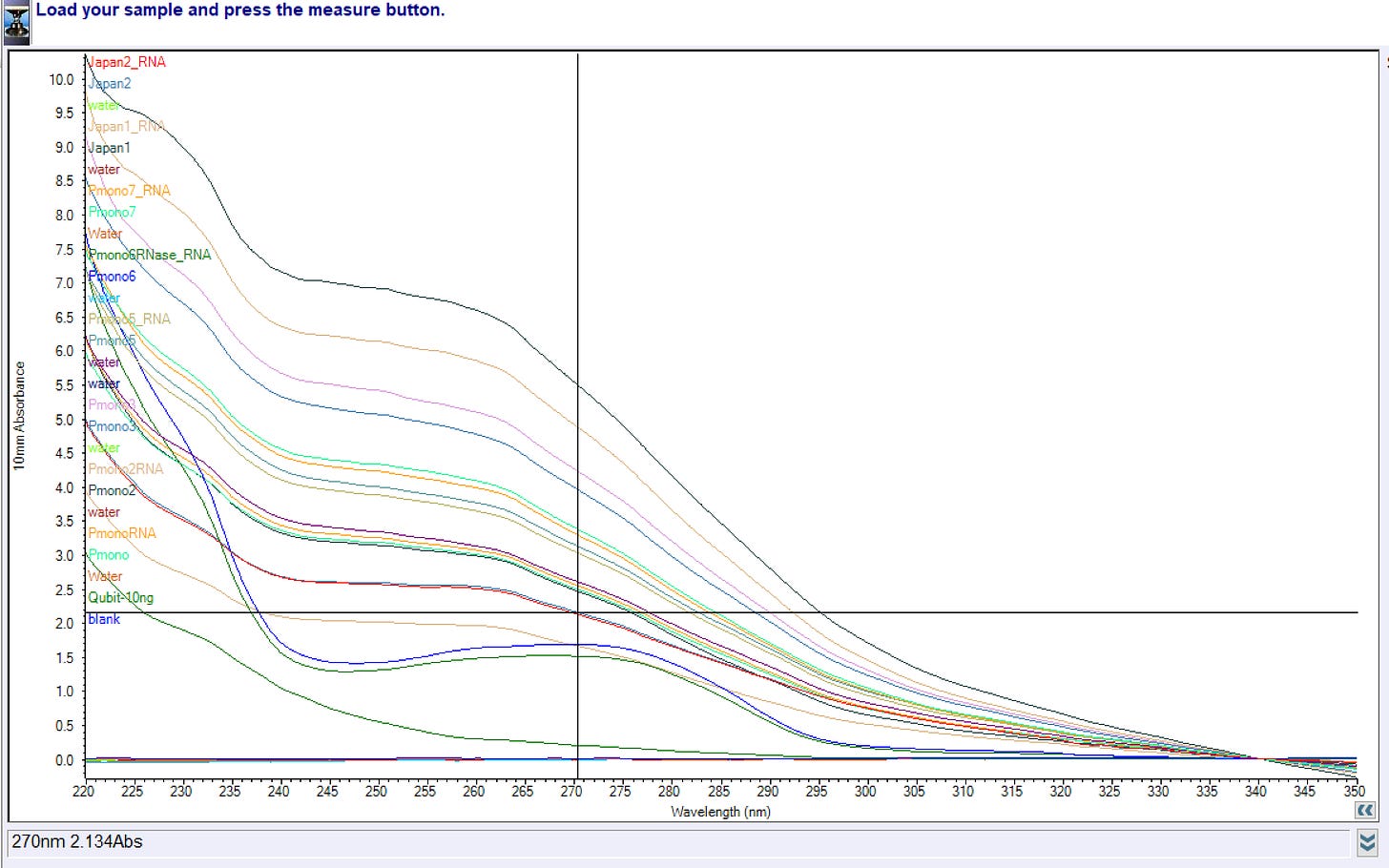
Fair point. Our purification of the DNA/RNA from the vaccine likely lost a lot of the material so lets remeasure this on 1ul of the vaccine itself.
Holy Shit!
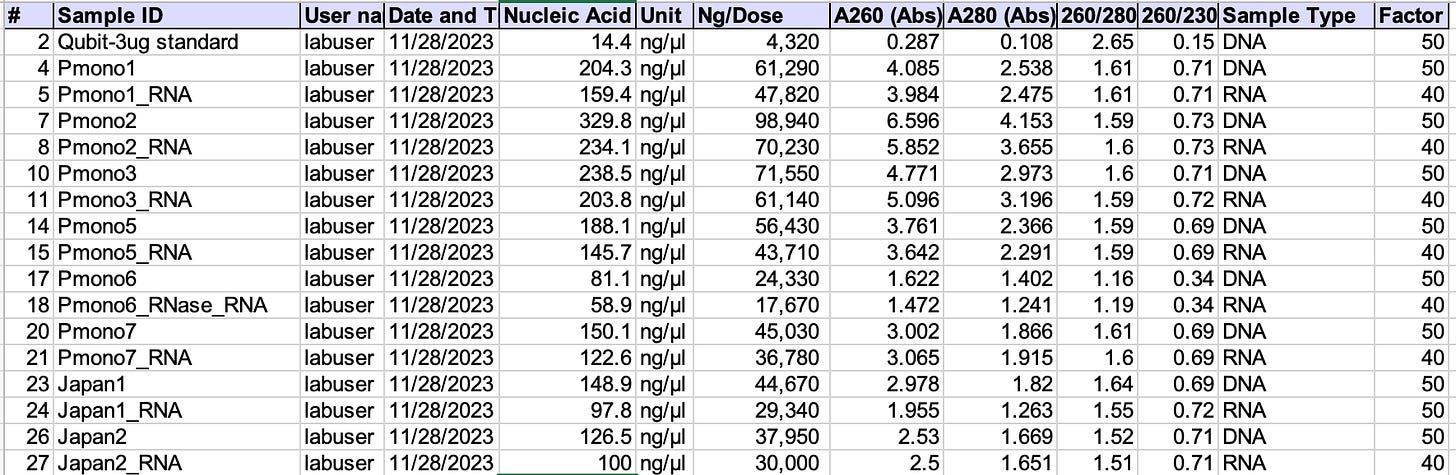
The 3ug standard (300ul of 10ng/ul) used for the Qubit is on the left. Comes in at 4.3ug. Measure it 5 times on the Nanodrop and you’ll see it hover with a 15-20% variance around this number.
The Pfizer monovalent vaccines have 30-99ug of RNA+DNA per dose. They should have 30ug total.
Now look at Pmono6. That sample is RNaseA treated and then CTAB/SPRI purified. No RNA. Just DNA. 17.6-24.3ug/dose. Not Nanograms per dose. 1000X higher than the FDA limit of 10ng/dose. Keep in mind, this sample was CTAB purified after RNaseA treatment as the RNaseA protein can obscure the UV spectra.
TritonX also obscures the UV spectra so it’s not easy to dissolve the LNPs to access the nucleic acids with enzymes like RNaseA without running a DNA/RNA purification step afterwords. This DNA prep can loose some of the smaller fragments of DNA.
So why didn’t we scream the largest numbers we could find back in March?
Because we’re honest.
We don’t have money in the outcome of these results. In fact, if we are wrong, we may be sued like Poland. When they are wrong the tax payer picks up the bill. This asymmetry of liability has made our reporting conservative regarding the issue at hand.
We also don’t think the techniques Pfizer is using to monitor these are legitimate. UV Spec is notorious in the genomics space for over quants of nucleic acids and we feel refuting their nonsense should never make the same mistakes they employ.
When a business wants to change the methods they are measured by every time you turn away, (RiboGreen for RNA, qPCR for DNA, UV spec for final product) you are dealing with scam artists and I doubt the regulators are this dim. I’m beginning to think they know this is a farce but that the public will never figure it out, or by the time they do, they will have a cozy job at Pfizer.
Reminder: If you want to measure RNA and DNA with the same yard stick; qPCR vs RT-qPCR is your best bet. They had the qPCR spike primers. They likely ran the RT-qPCR on their ‘RNA’ products and didn’t like the answer. So they scrambled to find any other tool that could give them an answer that would navigate the regs. Even under this ‘choose your own adventure in assay land’, they still couldn’t pass and we have reams of EMA communications with Pfizer showing they fail long into 2022.
While UV Spec is fast and easy to run, there is a limitation to the information it provides. Common lab reagents like TritonX can inflate the scores. The use of RNaseA or DNaseI doesnt reduce the UV Spec signal as you can see with Fluorometry and qPCR. It is the least specific QC they can use and its the last QC they do.
If you are new to this substack, there are previous articles posted on this topic that may help explain this.













Hello Kevin, I am a new subscriber so my first time here in the comments. As an analytical chemist with 40 years industrial experience, I can vouch that you are 100% right on. UV-Vis is a very non-specific method of analysis, meaning that about the only thing it's good for is quantifying an analyte that you already know is pure.. and you just want to know the concentration. The Gold standard for specificity might be something like LC-MS, whereby you first separate and deconvolute the various molecular species using liquid chromatography, followed by the use of mass spectroscopy to identify the molecular structure (and hopefully the identity) of the eluting fractions. Even that may not be enough if the fractions don't separate well enough in the LC... Using UV-Vis for QC on these transfection products is literally a joke, and they know it. Reason 842 why I am glad that I refused the transfections.
Thank you. I get the drift but I don't understand except for a very broad stroke. I thank you again and again.