Why is DNaseI failing
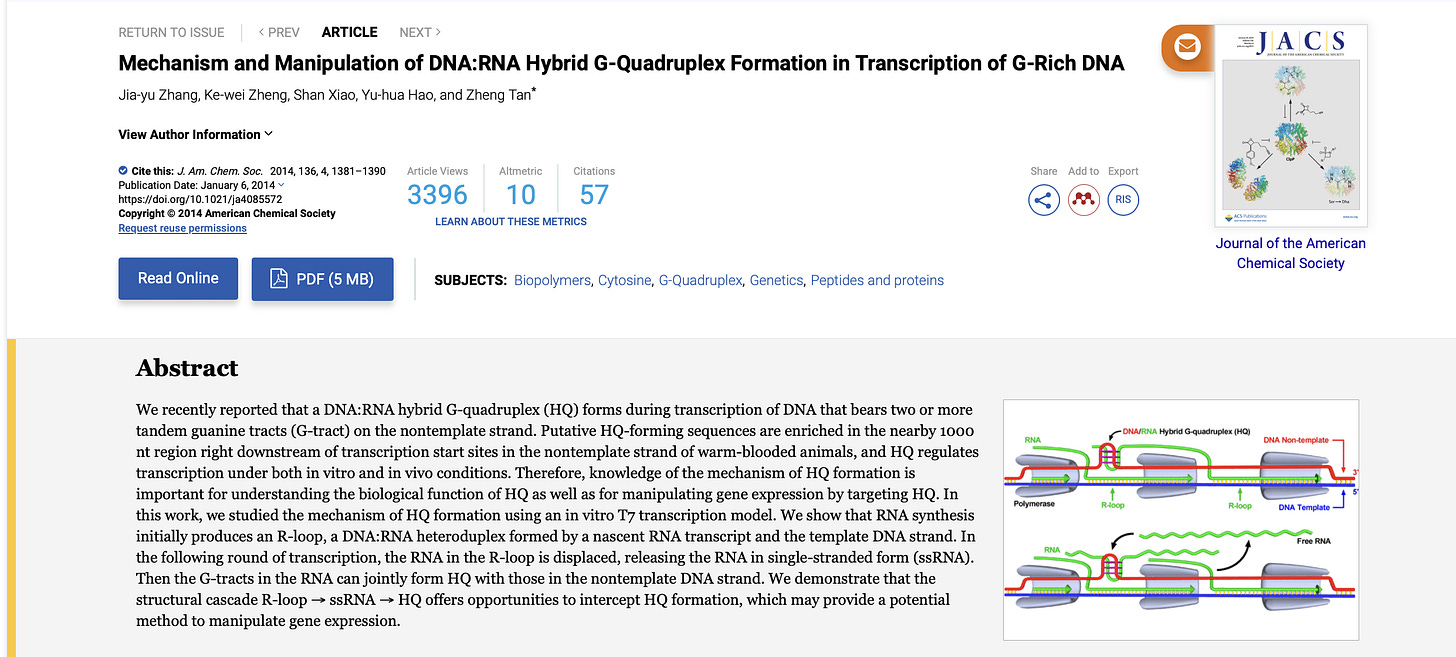
DNA:RNA hybrids
People often ask why is a simple DNaseI reaction failing for Pfizer? What other artifacts are present in this manufacturing process?
Sutton et al is a good place to start. Even in 1997 they knew DNA:RNA hybrids were treated very differently by DNaseI. The Pfizer vaccine is a DNA:modRNA hybrid and is in even more uncharted water.
Basically, when they use T7 RNA polymerase to copy RNA off of a DNA template, the resulting product is a DNA:RNA hybrid. A triple helix that DNaseI doesn’t recognize.
Some Moderna lots, while lower in DNA contamination show a 6 CT difference between the Ori qPCR vs the Spike qPCR. They are 100 fold better at removing the vector DNA vs the Spike DNA. This is likely the result of the Spike RNA being hybridized to its reverse complement DNA template that generated it and thus interfering with the DNaseI that is trying to remove the DNA. This RNA doesn’t exist over the plasmid ori sequence and hence it is DNaseI labile.
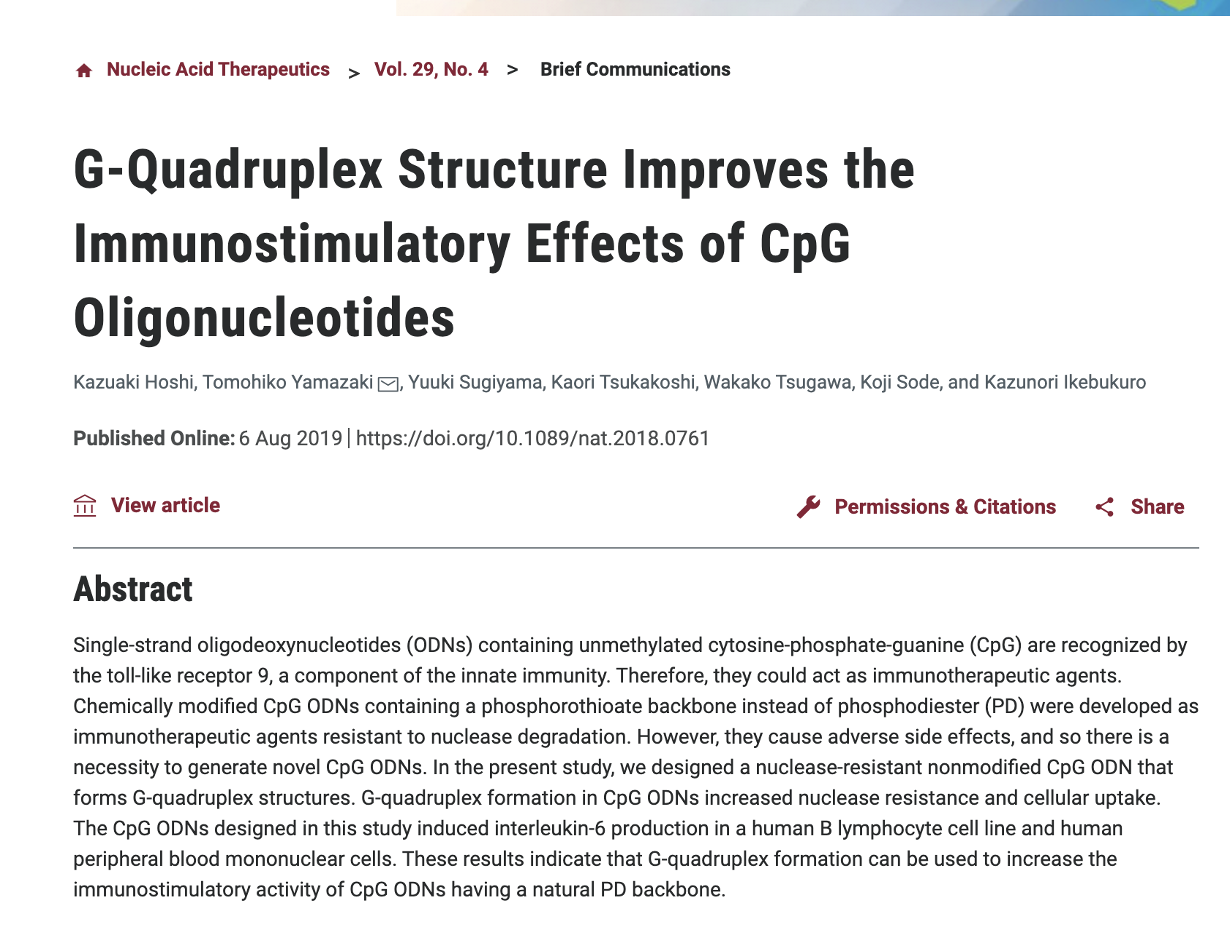
There is a previous substack on R-Loops but it is worth reiterating that Quadruplex Gs play a big role in regulating R-Loops.
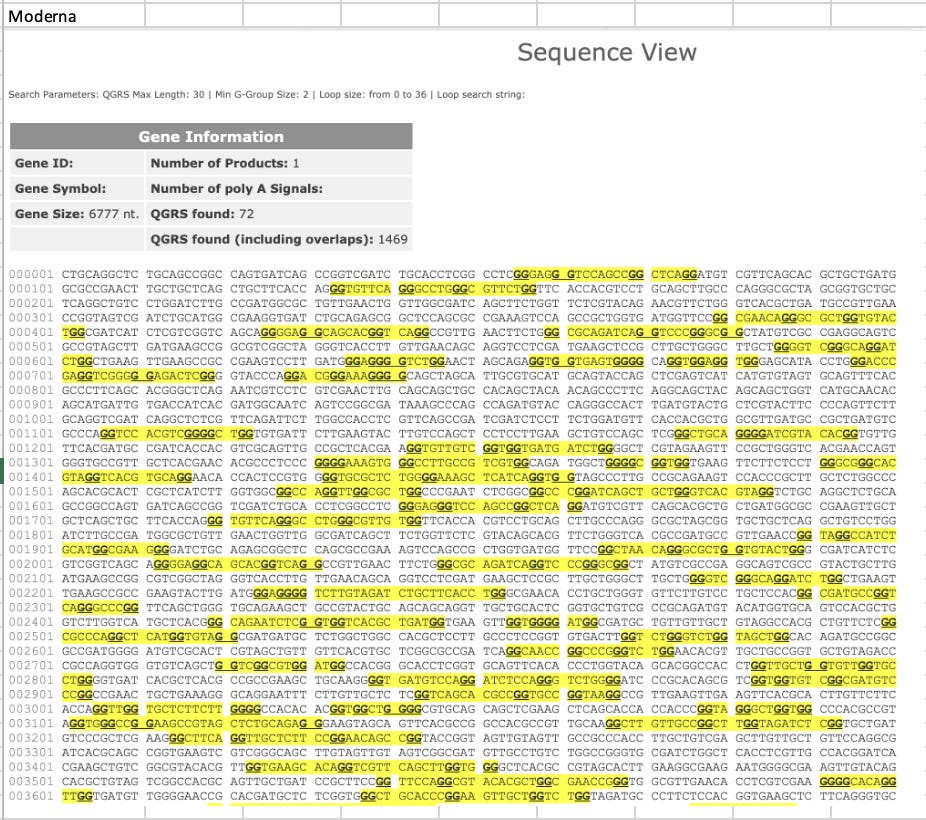
And the codon optimizations used for Pfizer and Moderna introduced many more Quadruplex Gs. We reported this in a PrePrint in October 2021, which had a hilarious censorship campaign imposed upon it covered by Tamara Ugolini.
At the World Council for Health, Byram Bridle ran through a few papers that touched on the GC content of the plasmids and their impact on integration.
Image from Byram Bridle presentation on dsDNA contamination-
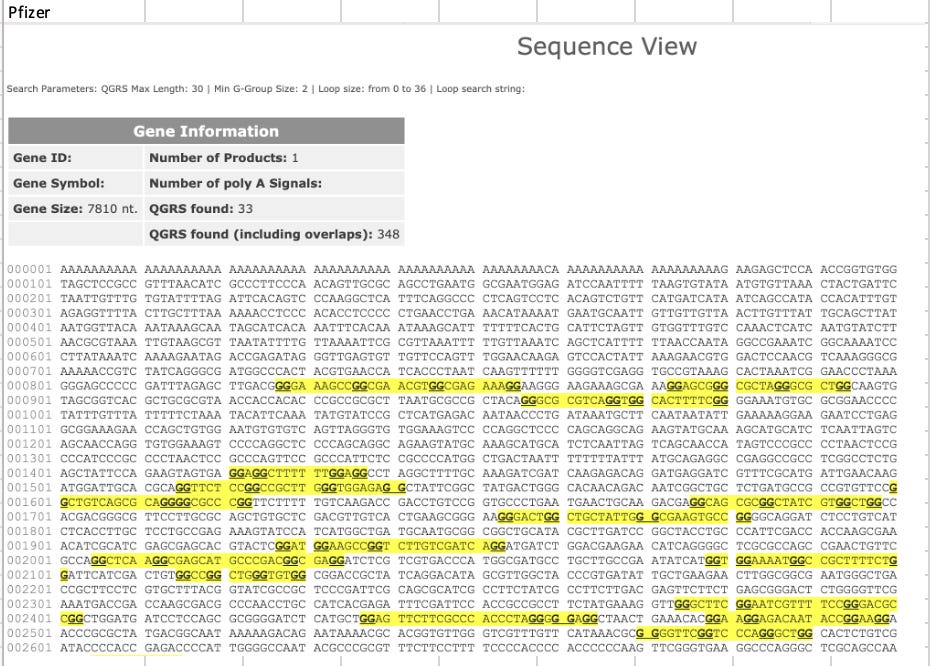
This led me to run a few calculations on the GC content and Quadruplex G content of the entire plasmid sequences. In the past, we just focused on the Quad Gs/GC content in the 4.2kb modRNA but now we know there is 7.8kb plasmid in the vaccine as well and these may have their own sequence motifs that need to be evaluated.
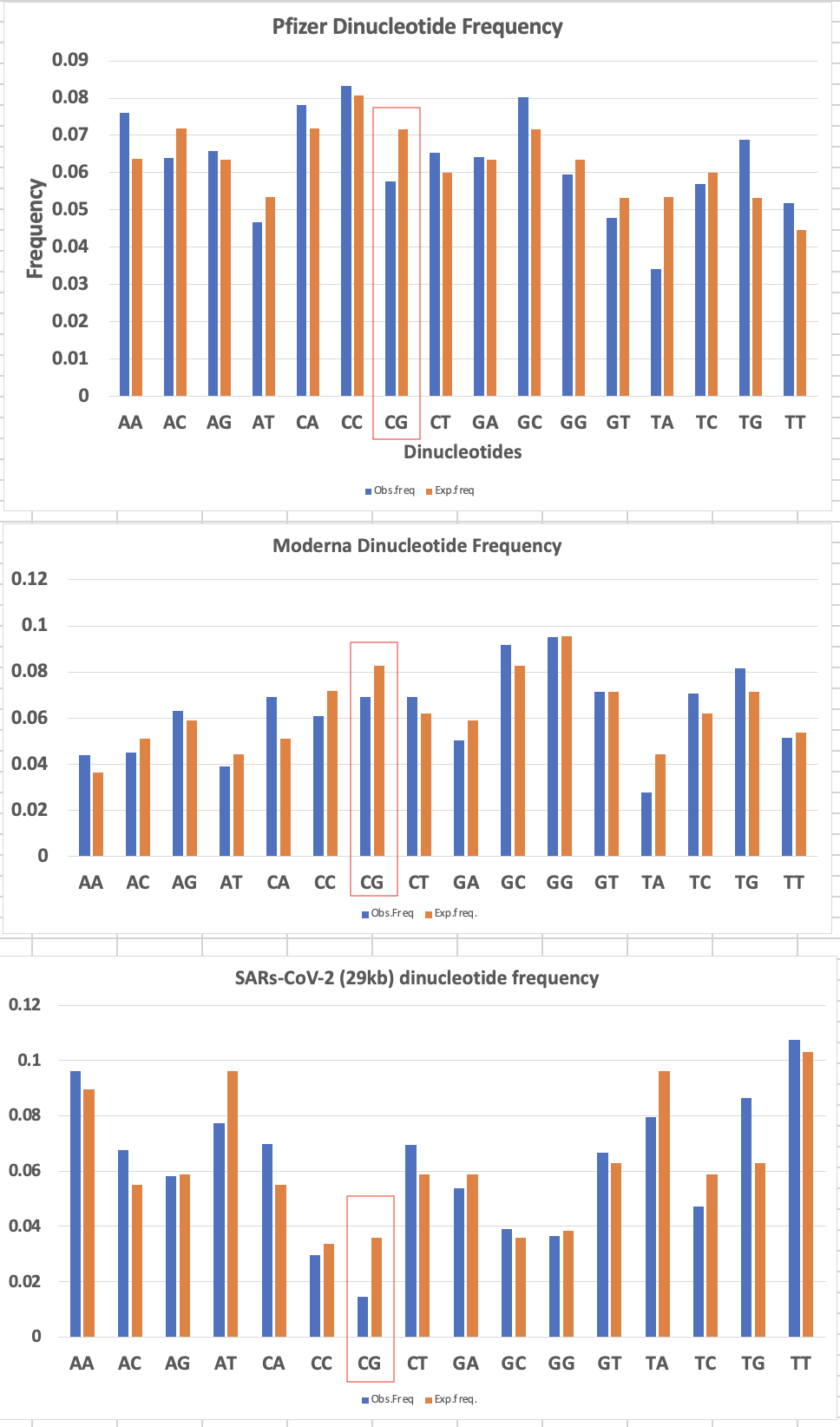
So did the codon optimization preserve the CpG we see in the virus.
Nope.
Heavily diverged. The virus avoids these and the vaccines and plasmids enrich for them.
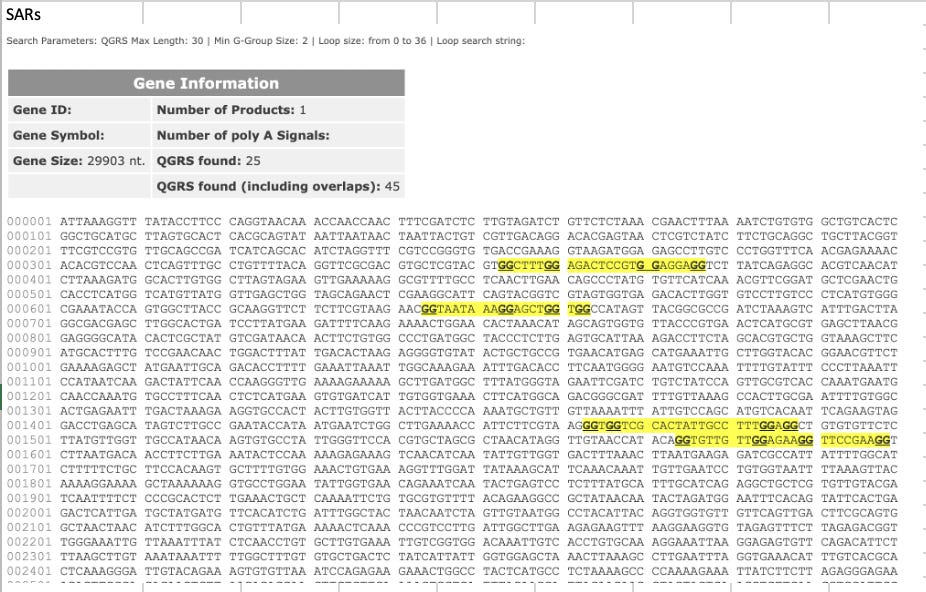
Another interesting artifact we see with DNA:RNA hybrids is in Phillip Buckhaults Oxford Nanopore Tech (ONT) sequencing data.
You’ll notice there is Half the Sequence coverage over the Spike regions than the Vector region? Why is that?
ONT uses a T4 DNA ligase to ligate their sequencing primers onto DNA prior to sequencing. T4 DNA ligase wants clean dsDNA to ligate primers to. It is not going to ligate to a DNA:RNA hybrid template.
In conclusion, the codon optimization and the use of modified bases that increase the secondary structure and melting temperature of RNA are likely making this DNA hard to digest. Monitoring only one region of the plasmid with qPCR , like the unprotected vector, might lead you to believe you’ve removed more DNA than you think.
Its important to understand the methods being used to measure this contaminant as there are nuances here that may not be obvious to regulators.
You’ll understand more of why this matters in the next post.
Stay Tuned.




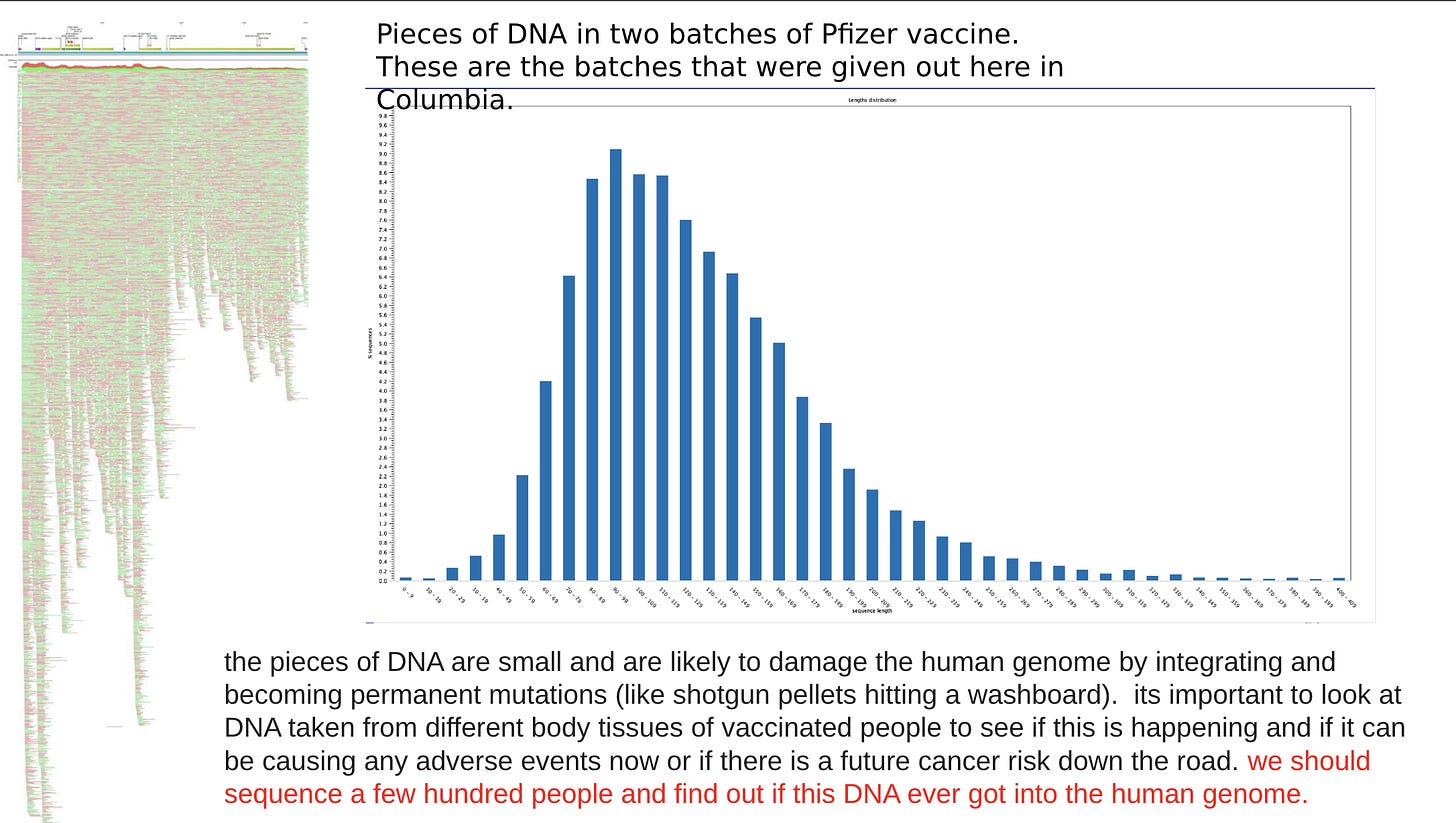


I'll probably have more than one comment but, right off the top...what is Steve Kirsch referring to when he said he would have to sacrifice his deltoid muscle if he were to volunteer to for DNA-integration testing? Is that true?
****
THIS is what Steve wrote:
"I volunteered for a full gene sequencing study, but they said they’d have to cut off my deltoid muscle, so I changed my mind."
Please do not assume I need a be educated on what a biopsy is. FFS my husband has cancer, right now. ^What Steve wrote sounds nuts, tbf.
As soon as the sheeple around us begin to “intentionally” educate themselves with the same effort they keep themselves ignorant—blindly-accepting vaccines & war—the sooner the entire world can pull it’s head out of it’s ass and save itself.
Again, this all began prior to 1960, but it was in that decade Coronavirus and the mRNA (spike-protein, gene-altering, militarized nanotechnology) Vaxx Protocols were developed—side-by-side—to “terrify” Animal Farms (sheeple) to accept a worldwide, multi-faceted Lab-Rat experiment.
None of this is about saving humanity, but to use people to death to better the health of billionaires & trillionaires with bomb shelters. If you’ve taken the time to read my post to this point, then I fu🤬ng BEG you to click on this link to a video David Nixon just posted, then forward to every blind-bot you know: https://substack.com/@jeffreyplubina/note/c-42176817?utm_source=notes-share-action&r=1qpmjb