Plasmid DNA replication in BNT162b2 vaccinated cell lines
FLCCC interview & more
I had the pleasure of speaking with Dr. Pierre Kory and Dr. Paul Marik last week.
(
on Twitter @drpaulmarik1 )I got side tracked on bit too much of the template switching and frame shifting background to get into the DNA integration problems but Dr. Hiroshi Arakawa downloaded our integration data and performed a deeper analysis.
You may need a translate option on your browser but this link has instructions on how to do that straight from the X browser.
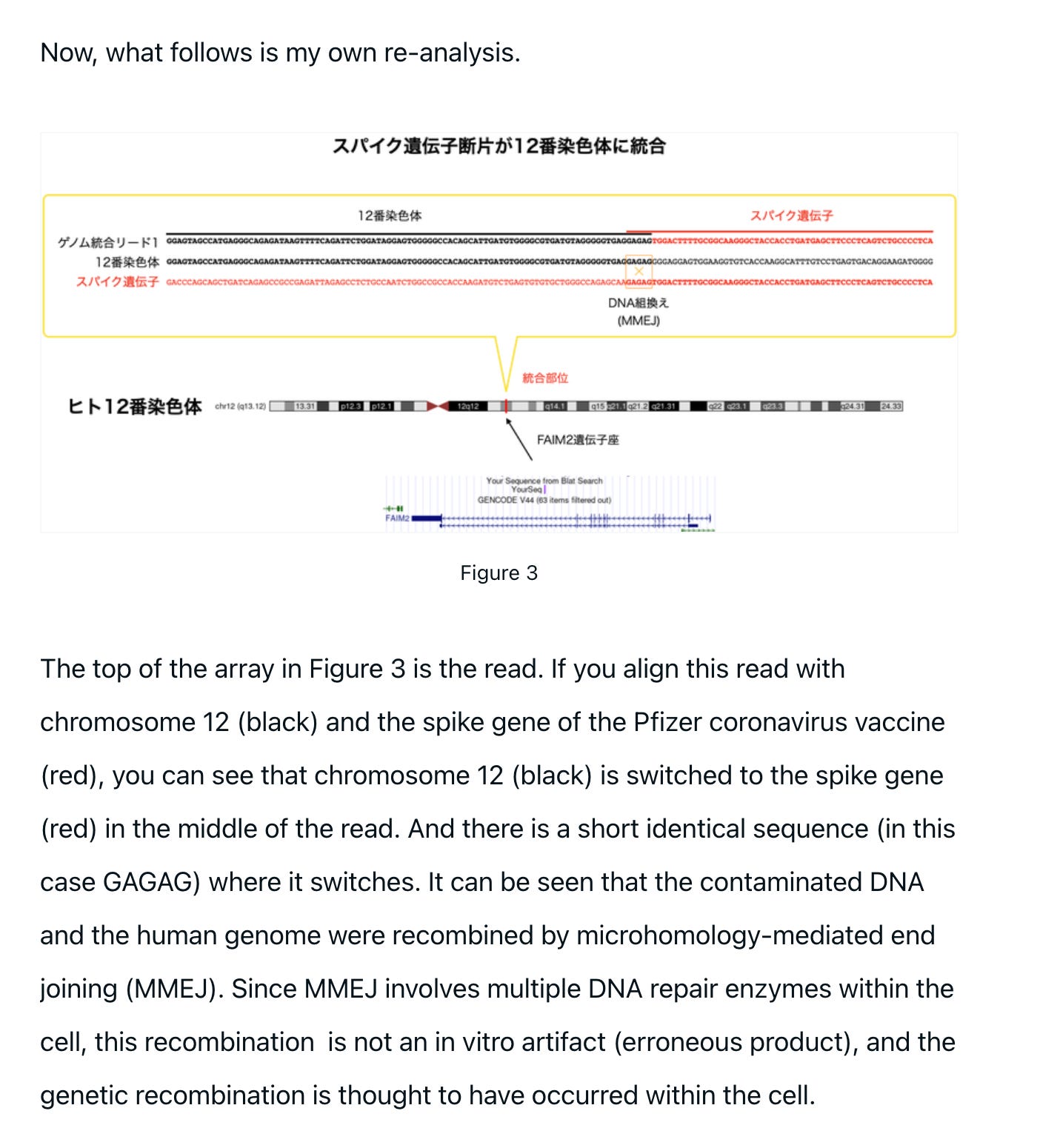
The integration on Chromosome12 has some hallmarks of micro-homology mediated end joining (MMEJ).
Break-
Break-
The article also suggests the chromosome 9 integration events needs further scrutiny. We used the DRAGEN mapper and the reads we published were unmodified to ensure our reads files had the identical hash as the sequence provided to us from the sequencer. Any read trimming would alter this hash and break the data audit trail. The Chromosome 9 integration even may be influenced by this but the Chromosome 12 event survived read trimming.
This is why we put the reads public immediately even though we knew the data was preliminary and in need of further verification. The best peer review is performed live and in public.
The Chromosome 12 integration is looking more real after Dr. Arakawa’s analysis and the Chromosome 9 integration has weaker evidence.
This leads me to a very important finding that I hope others in the community will also pay close attention to.
Plasmid DNA replication in BNT162b2 treated OvCar3 cells
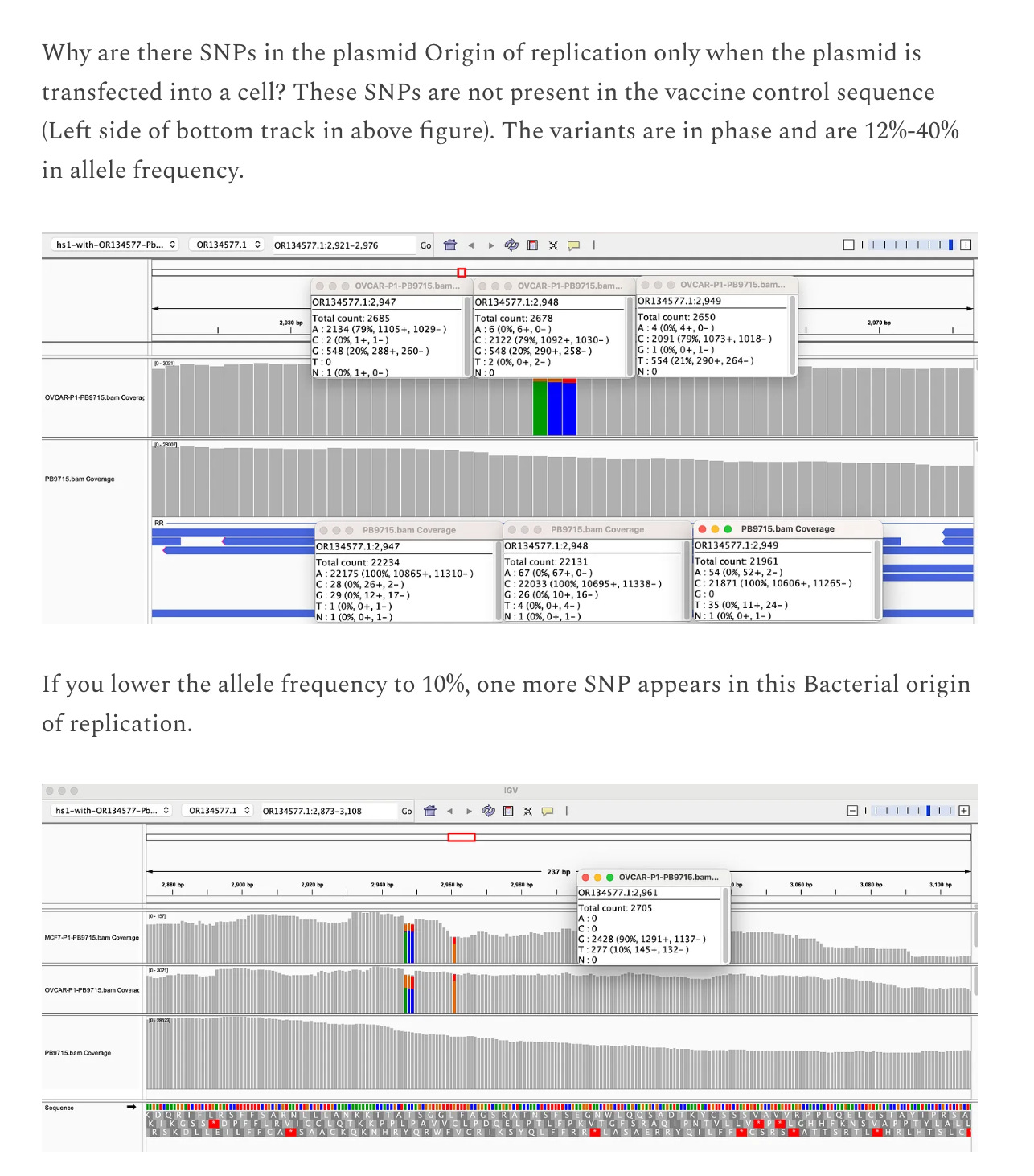
The vaccine DNA in the OvCar3 cells is different than the vaccine DNA in the vaccine before it was administered. There are 5 SNPs found in the plasmid DNA (above 10% allele frequency) once it is in the cells.
Of note the Sequence coverage of the vaccine in the cells is 3000X. The cells are sequenced to 30X sequence coverage.
3000X means every base in the plasmid is sequenced by 3,000 independent DNA molecules while the cells it is in are only sequenced 30 times. This implies 100 copies of the plasmid compared to the genome. This plasmid DNA is likely fragmented but in high copy per cell. I was not expecting this much contaminating DNA from the vaccines.
Some of this is described in more detail on the end of this integration substack.
Vaccine targeted qPCR of Cancer Cell Lines treated with BNT162b2
Ulrike Kaemmerer has treated MCF7 and OvCar3 cancer cell lines with various vaccines. Once transfected they performed cell passaging on these transfected cell lines to dilute out the residual vaccine and identify cells which were transfected. They performed Immunohistochemistry (IHC) on these cells and documented spike expression levels.
Below is an IGV coverage map of the reads from the vaccine in OvCar3 (Top Track). The grey histogram is the coverage at each base. The vertical colored lines are the SNPs in the sequence. We expect SNPs in the spike region as the reference genome was a consensus that was built from a Bivalent Pfizer vaccine and we are sequencing a bivalent Pfizer vaccine. So we expect certain bases in the spike region to have 50:50 alleles that represent the sequence differences between the Wuhan1 and Omicron sequences.
We do not expect SNPs in the plasmid backbone unless the cells are replicating those sequences and making errors in the process.
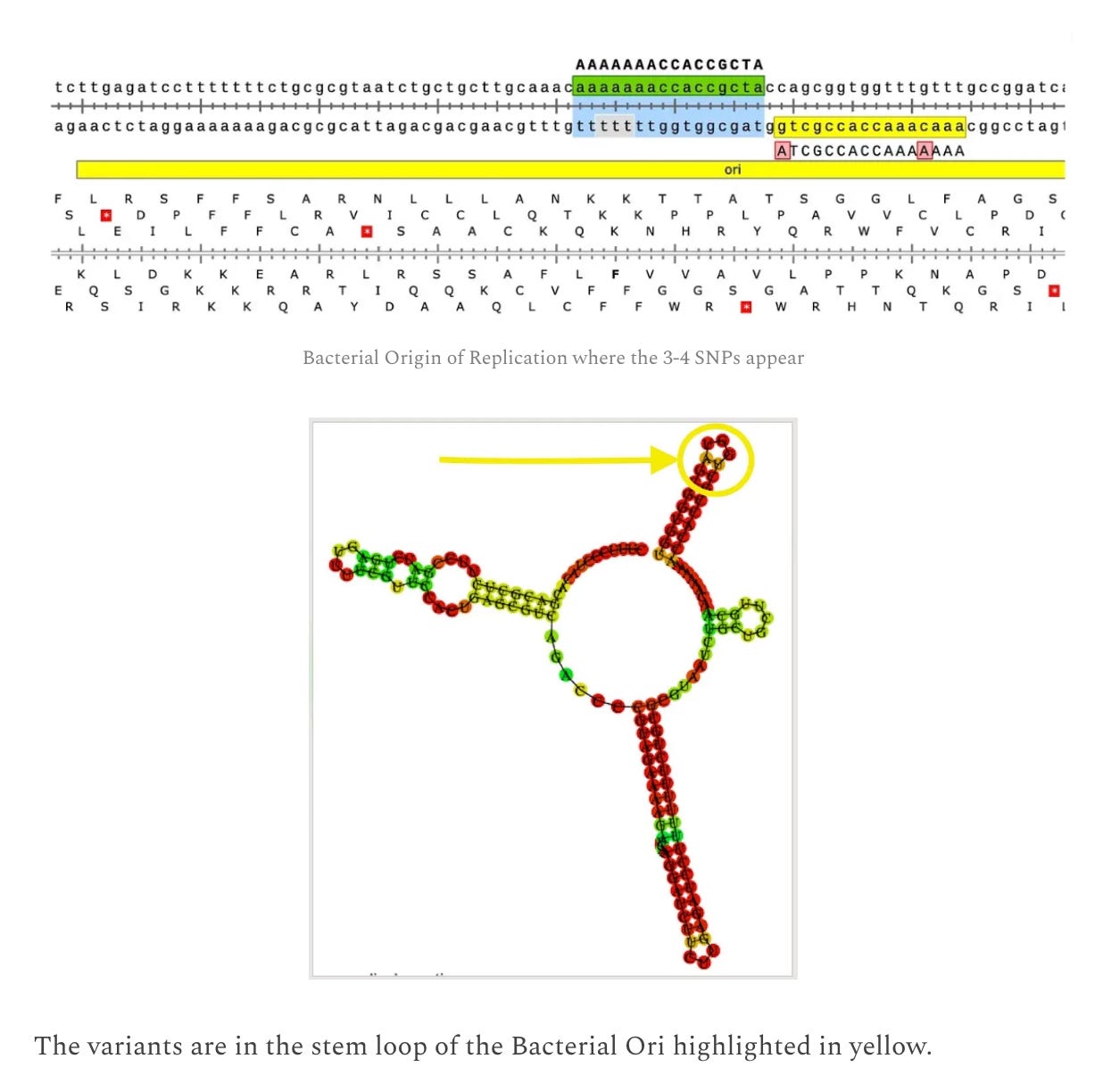
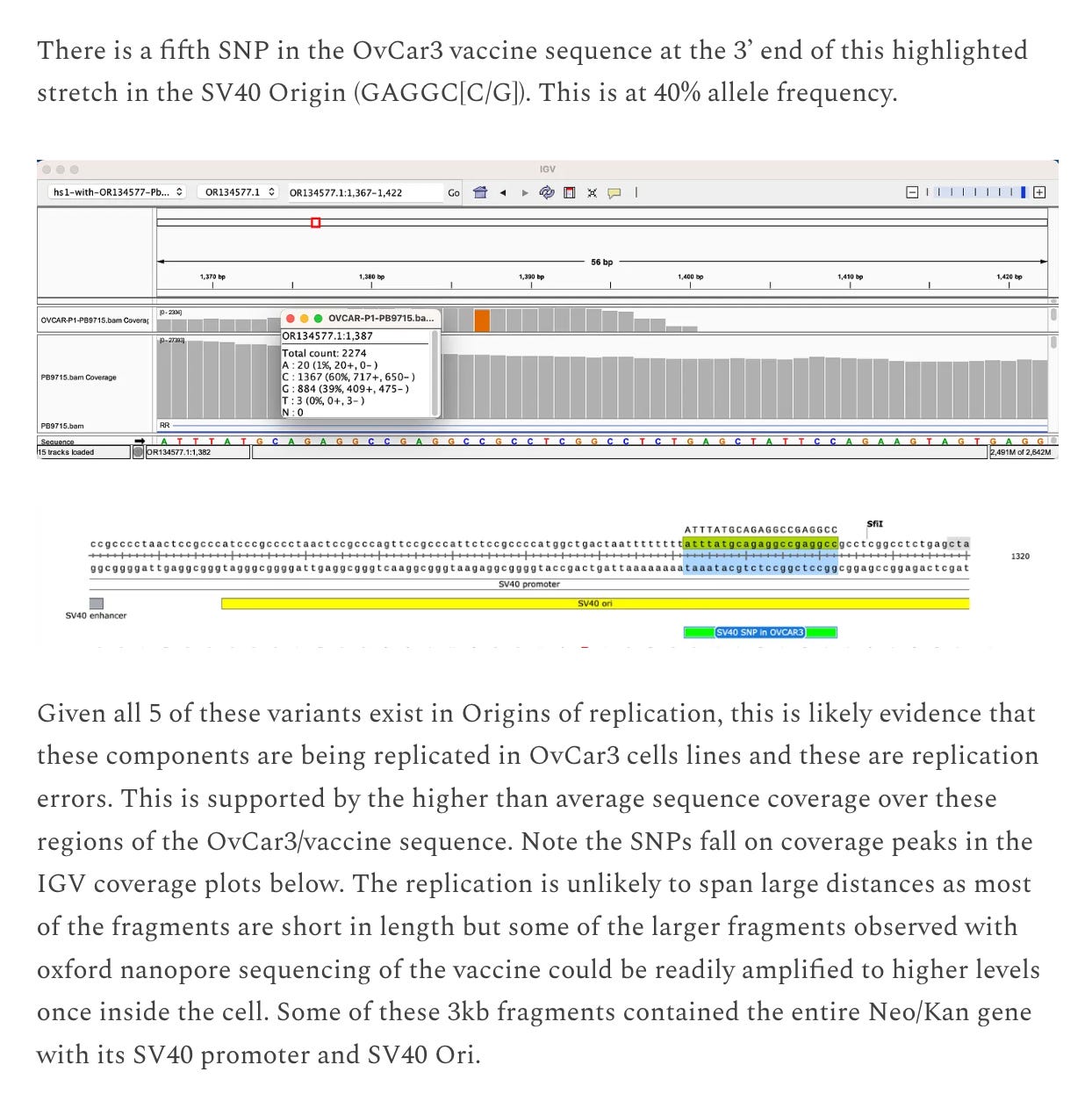
The 5 SNPs we do see in the vaccine DNA are in the F1 Origin of replication, bacterial origin of replication and the SV40 Origin of replication. All 3 regions regions have more sequence coverage implying the DNA is being replicated in the cells and we are witnessing DNA replication errors manifest in the form of SNPs in the origins of replication.

This is an important issue considering the SV40 origin of replication was omitted when submitted to the regulators. The regulators are claiming this DNA isn’t functional and we have hard proof that it is replication competent in OvCar3 cell lines.
This is also a reminder of the naivety of regulations that don’t consider the type of DNA that is contaminating the vaccine. 10ng of host cell DNA is very different than 10ng of 7kb gene-therapy plasmids that contain mammalian origins of replication, promoters, enhancers and aminoglycoside resistance genes.
Small amounts of contamination can be amplified inside the cell making the current DNA regulation loop hole large enough to drive a truck through.
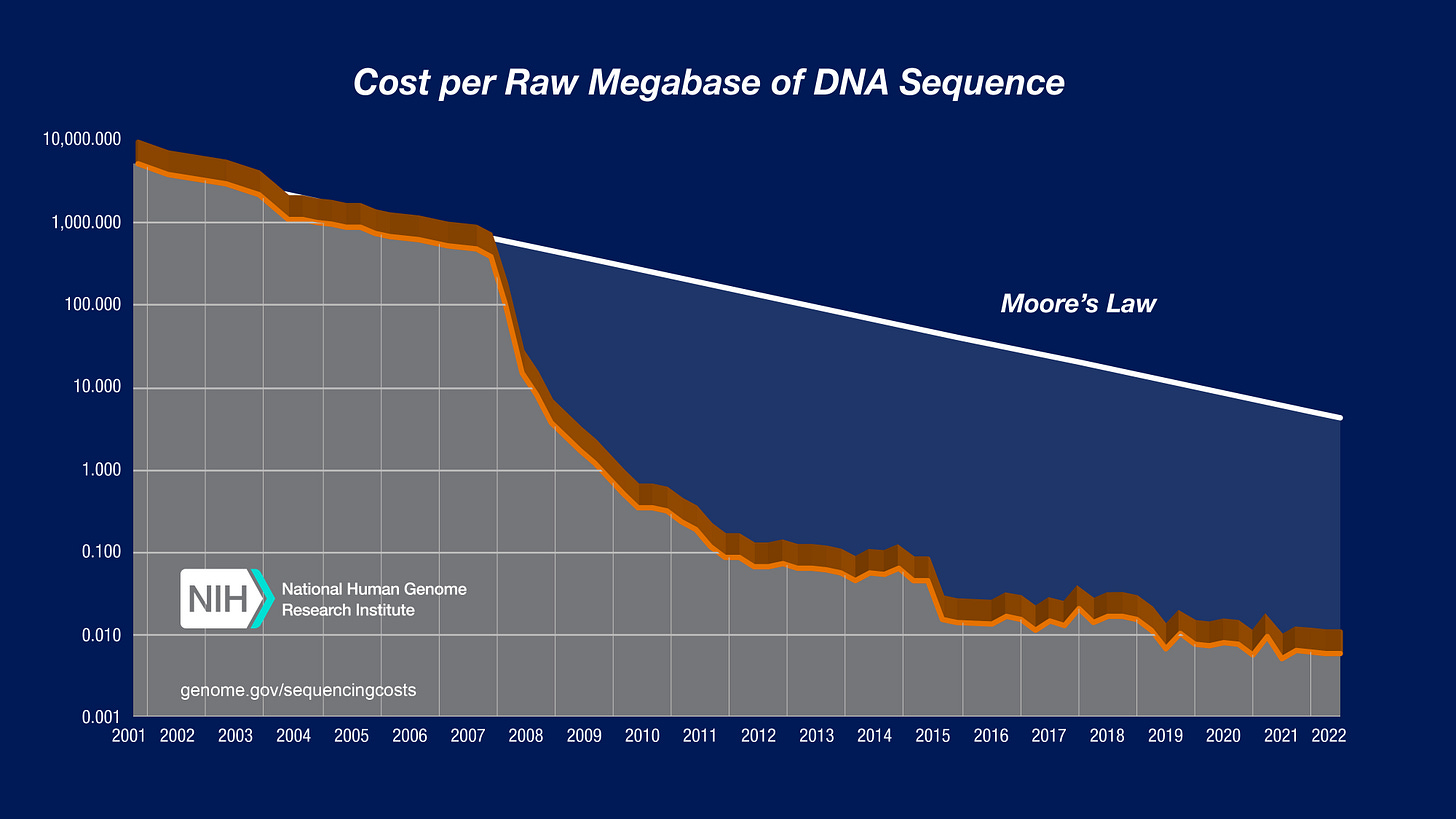
The cost of DNA sequencing recently plummeted 100,000 over the same time period the FDA relaxed the vaccine DNA contamination limits 1000 fold (10pg to 10ng). These regulations are DNA blind. They don’t care which DNA is contaminating the vaccine despite how easy it is to know precisely how much and what kind of DNA contaminates each vaccine.
It baffles me that sequencing isn’t performed on every vaccine lot but I guess that’s what you get when unelected officials can hand out liability waivers and mandates to the vaccine manufactures.
While the EMA has many metrics these vaccines must meet, most of these measurements are “trust me bro” measurements performed by the manufacturers with results handed to the regulators.
They would be wise to remeasure the risks in the vaccine that are amplifiable or exponential risks described in this short video.
I discussed this briefly at the ICS5 meeting organized by
For those just here for the cat pictures: That is Ollie scouting a black bird in the window while listening to Black Bird from Fat Freddy’s Drop.
Lyrics seem fitting-
Where there was one before
Now there are many
And as the days go by
I see them multiply
I can't eat
I can't sleep
Because I know they'll be watching me
Descending on the breeze
Their shadows keep falling down on me
There's a blackbird
In my garden
Oh!
There's a blackbird
In my garden
Oh!









Kevin, your SS's tell the story and record the history of this Crime Against Humanity.
Thank you for bringing together the recent news and putting it here. Very helpful source for friends who are still getting up "Fool's Hill".
Great article as usual. After all these integration risks there is a bonus.
Because of frame shifting who knows what proteins and antigens are actually being created!
Clearly, Big Pharma has no idea or does not want to tell us.