SV40 Enhancer qPCR
A tool for investigating long vax and persistent spike duration.
Given it is now known that the SV40 enhancer in the Pfizer vaccine is a potent nuclear localization signal (NLS) described by Dean et al., it seems prudent to design an SV40 qPCR assay. If there is ever going to be a dsDNA induced genome integration event its most likely going to entail the NLS given the dsDNA in the vaccines is mostly fragmented.
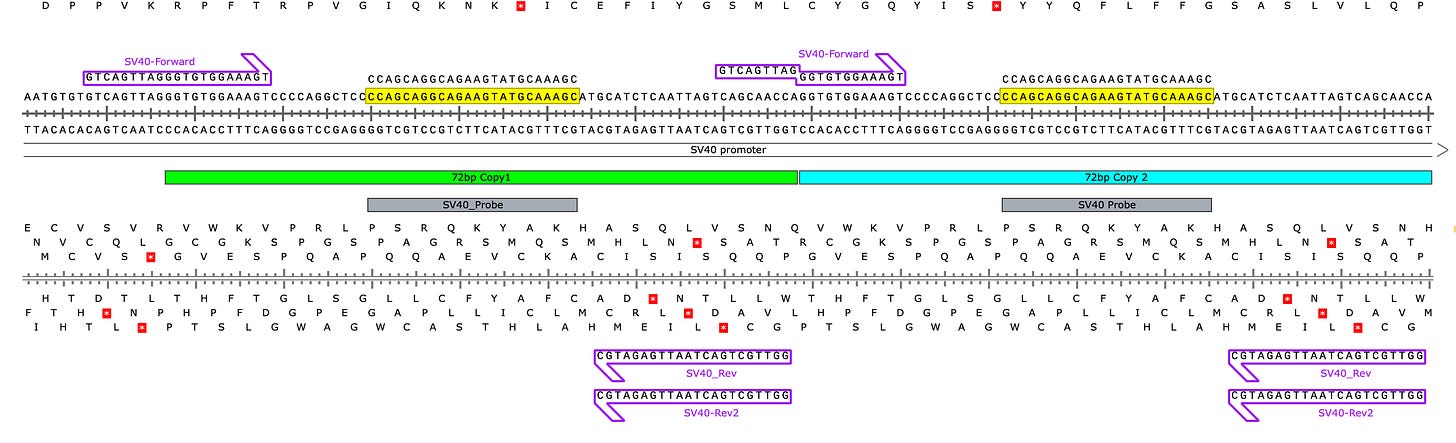
To do this, we targeted the 72bp tandem repeat (also known as the bidirectional Enhancer element) in the Pfizer vaccine. This assay will not work on the Moderna vaccines as they do not contain this sequence.
This assay may be helpful to track which tissues may have SV40 elements from the Pfizer vaccine still present.
The assay is designed to be more sensitive than the Spike and Ori assays as it can take advantage of the 2 copies of the sequence in the vaccine. We have a Forward primer that bridges the left junction of the SV40 enhancer to the plasmid but doesn’t fully bridge the 72bp-72bp union (Green-Blue bar in Figure 1). We have a probe that lands in 2 places (Yellow in Figure 1) and Reverse primers than land in two places (purple arrows pointing left- Figure 1).
This assay was put into the Texas Red channel so it will fluoresce in a different channel than the Spike and Ori assays and thus is can be readily multiplexed into the existing MGC assay being used to survey the vaccine vials.
Below (Figure 2) is our first attempt at this. SV40 amplification (Gold) is more sensitive than the Ori (Green) and Spike (Blue assay). This is likely the result of their being two places for the Probe and the Reverse primer sites to amplify (Figure 1).
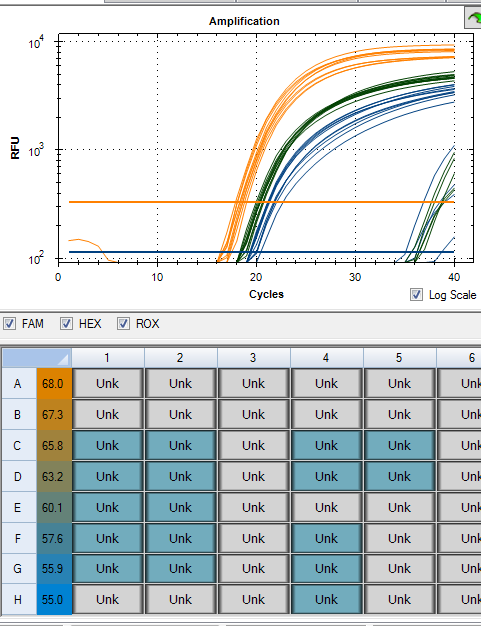
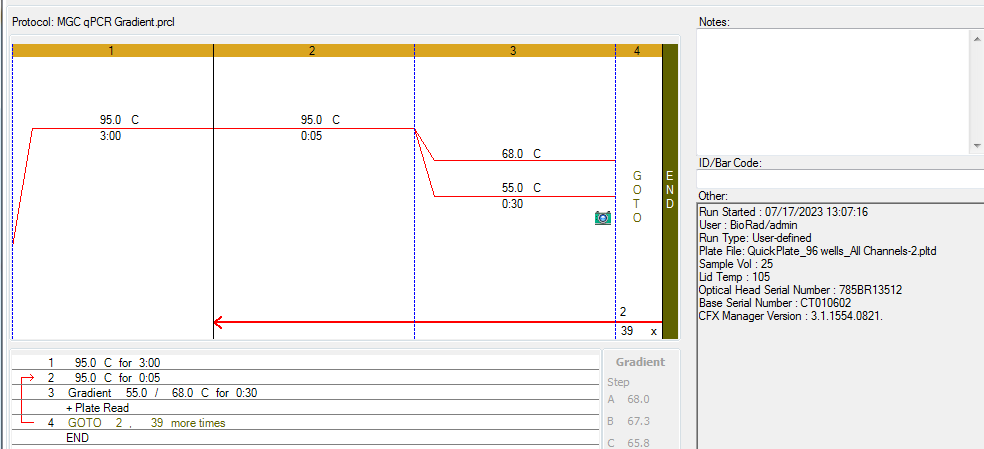
The conditions used for this amplification explored a temperature gradient (55°C-68°C) and 65°C will continue to work for all 3 assays.
Mastermix used Medicinal Genomics qPCR kits as previously described.
246µl ddH20
10µl Spike Primer-Probe
10µl Ori Primer-Probe
10µl SV40 Primer-Probe
114µl MGC Master Mix (Green Cap)
24µl PCR Buffer (Blue Cap)
13.8µl Master mix/ well
5µl sample (2µl Vax/ 3µl ddH20)
This assay may be helpful in screening clinical (Long Vax) samples for PCR positivity for Spike, Ori and SV40 Enhancers. Positive samples should be prioritized for whole genome sequencing or TagMap Illumina sequencing as described by Zhang et al.

Thank you Kevin. The truth will prevail. We are fortunate that you are doing this work. Peace.
I see this as a major upgrade. I wonder if it would be sensitive enough to detect the vaccine contaminants in the blood of recently-vaccinated patients (although many of them would probably rather not know).