More Holes in the Achs paper
Their CE methods are designed to find nothing
Having been the Team Leader for R&D on the Human Genome Project at Whitehead/MIT and managing a fleet of over 100 ABI Capillary Electrophoresis instruments, I naturally ignored the CE section of the Achs paper as I knew that would be a sham. Not because CE doesn’t work but because you have to feed them a large amount of pure DNA to see anything and you cannot feed them dirty preps. The old slab gel based 377 sequencers were very tolerant of dirty preps but once the HGP upgraded to use capillary electrophoresis, we had to change the entire prep pipeline. These systems use electrokinetic injection of the DNA and it needs to be in a pure water or formamide solution for this injection to work.
I finally got around to reviewing the Achs CE methods and I was shocked to find what an overt snow job this section of their paper is.
For a paper that drones on about their pristine validated methods it should shock everyone that they resorted to using an Agilent 5200 CE instrument that is NOT capable of detecting the DNA at the 10ng limit.
Here is the simple math.
FDA limit = 10ng.
Vaccines are 300-500ul.
10ng/300ul = 0.033ng/ul or 33pg/ul is what the vaccine is expected to be under!
So what do the specs of their instrument say?
50pg/ul is its lower limit of detection for a DNA smear. Optimal concentration is 20X this at 1ng/ul. DNA below 75bp is probably lower than this as the smaller DNA doesn’t stain as well.
Here are the methods from Achs et al
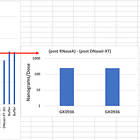
On top of this overt snow job of theirs, they didn’t perform a DNA prep for this. They simply added 10% Triton X-100 (not clear if this is final concentration or starting?) which will fowl up the electrokinetic injection.
On top of the unknown amount of Triton X-100, they also toss in an unknown amount of RNaseA. They claim 0.3ug/ml of RNaseA but nowhere in their methods do they disclose the total sample volume. Then they add 0.1xTE. So we have a LNP mayonnaise, PEG, salt, Triton X-100, RNaseA and TE soup they are trying to inject at 3.0Kv for 5 seconds into a machine not designed to detect 0.033ng/ul. Of course they can’t detect anything.
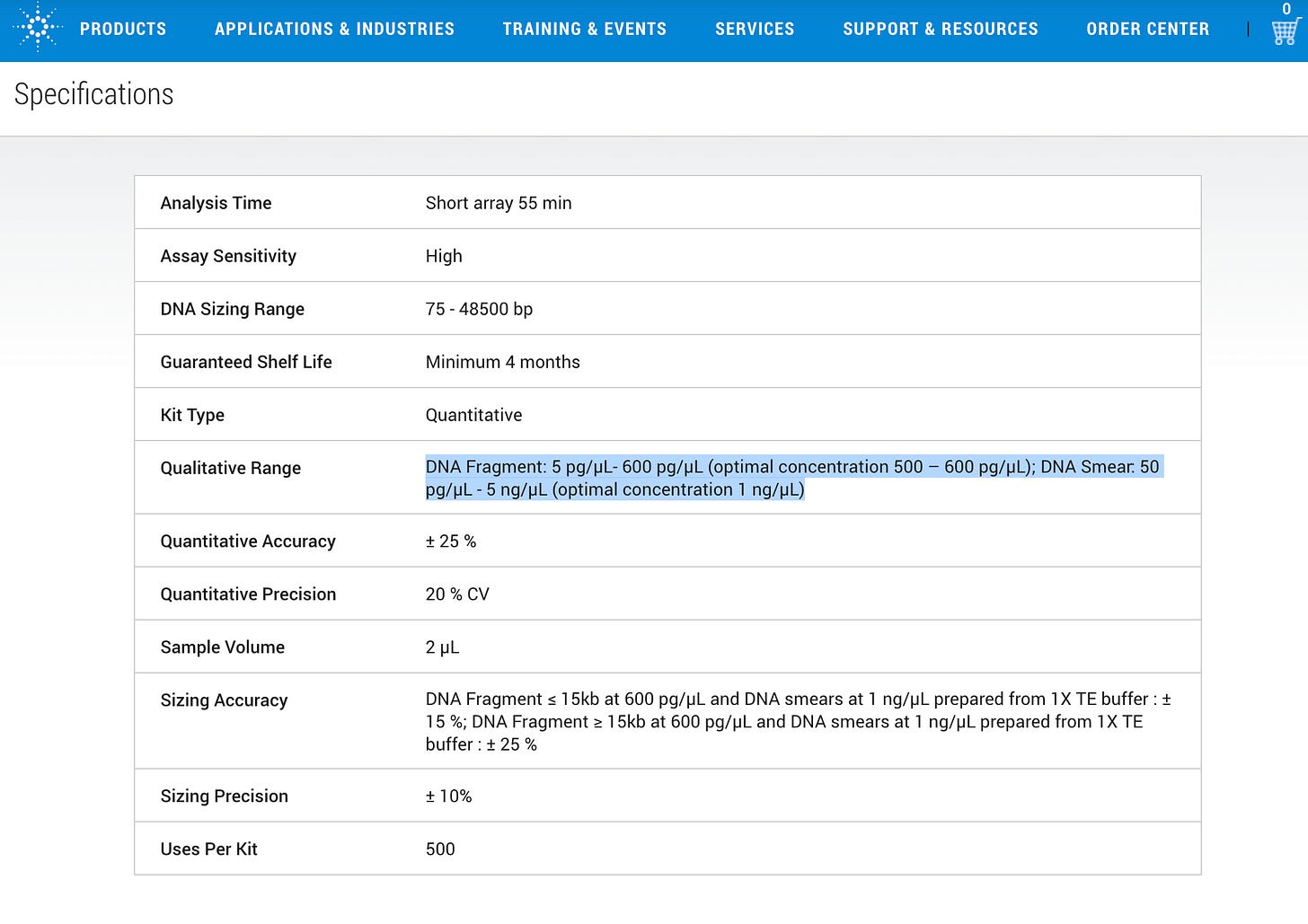
And just like their fluorometry parlor trick they attempt to show some DNA ladders injected into the CE machine but they are large pure plasmids/fragment ladders (not smears) and there is no LNP background matrix. Note the machine has 10X lower sensitivity for smears (50pg) vs ladders (5pg).
1)The 10ng limit is below the detection sensitivity of their instrument (Egg on Face moment)
2)They have zero matrix controls for the Electrokinetic injection.
Once again, The Ach’s paper can’t even pass a simple AI check which begs the question, why do we need Nature peer review?
Another example of their attempts to intentionally distort this picture.
Claude.ai-
Yes, the Agilent Fragment Analyzer systems do use electrokinetic injection for capillary electrophoresis.
Confirmed: Electrokinetic Injection
The Agilent validated methods use electrokinetic (voltage) injection, for which nucleic acid enters the capillaries without reducing the sample volume. An electrical current is then used to move the nucleic acids through the gel matrix.
Samples are voltage-injected into the capillaries and each sample moves through an individual capillary in a size-dependent manner.
Why This Matters for Achs et al.:
Electrokinetic injection has significant bias issues:
Charge-dependent injection: Highly charged molecules (like DNA) get preferentially injected over neutral/low-charge species
Size discrimination: Small fragments inject more efficiently than large fragments
Ionic strength effects: High salt samples show reduced injection efficiency
Matrix effects: Sample composition affects injection reproducibility
Critical Implications:
Since vaccine samples contain:
High salt concentrations (phosphate buffers, NaCl)
Protein/lipid matrices (affecting ionic strength)
Variable fragment sizes (contaminating DNA could be various sizes)
The electrokinetic injection would introduce systematic bias in the quantification, potentially:
Under-detecting large DNA fragments
Under-injecting from high-salt vaccine matrix
Inconsistent quantification across different sample types
This Adds Another Layer to the Detection Problem:
Not only is their detection limit too high (50 pg/μL vs 33 pg/μL regulatory limit), but electrokinetic injection bias could cause them to systematically under-quantify DNA in the vaccine matrix.
Proper methodology would require:
Hydrodynamic injection (pressure-based, no charge bias)
OR sample cleanup/dilution to reduce matrix effects
OR internal standards to correct for injection bias
This injection bias compounds the detection sensitivity problem, making their “below detection” results even less reliable for regulatory assessment.
/End Claude
Every section of the manuscript is invalid and it appears intentionally deceptive and designed to be such.
1)The qPCR primer designs are a mess. 63bp for KAN, 233bp for Spike. This hides the excessive Spike DNA seen in 4 other studies (McKernan, Speicher, Buckhaults, Fleming)
2)The equation for converting qPCR signal into nanograms doesn’t spell out the fragment lengths they use to compute the yield. No one can reproduce this unless the calculations are clearly spelled out.
3)The fluorometry introduces a biased DNA prep. Only done for Fluorometry method which look like intentional manipulation. Poor matrix controls in their standards. No adjustment for Georgiou et al
4)Illumina sequencing (ironically uses fluorometry to QC the DNA not qPCR) fails to represent the true fragment length. The authors have an Oxford Nanopore system but refuse to use it.
5)CE methods are mythical. Stated instrument doesn’t have the sensitivity required to find anything near the limit and they have no controls for dirty matrix injections.
6)They are a vaccine research institute with funding from an mRNA vaccine company but continue to declare no conflicts despite these being raised by comments on their preprint and in other preprints published.
Other reading