FDA White Oak Lab Finds 6X to 470X DNA contamination in mRNA vaccines
Excellent Work From High Schoolers
Fellow Anandamigo
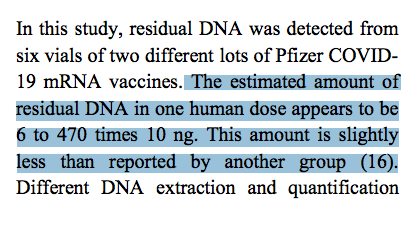
alerted me to a publication that came out in the Journal of High School Science that found 6-470 times the 10ng DNA FDA limit in Pfizer mRNA vaccines (Wang et al).You can find her Exclusive story on this here
It didn’t take more than a millisecond for the usual trolls to go ad hominem on the authors from Centreville High School in Virginia. These people will eat their own.
The proper approach in science is to ignore the authors, ignore the institutions, ignore all the social engineering bias than can result from such appeals to authority, and just read the methods and the results. Draw your own conclusions. Often the Abstract and Conclusions the authors provide are poetry to appease the gatekeepers at Journals.
These students worked closely with FDA scientists, used their lab, and appear to have performed the work with their technical support.
We are grateful for Drs. S. Liu, P. Selvaraj and
Wang of the FDA for technical support and
providing materials.
Now for the bombshell-
You read that correctly. Even in FDA labs, the few lots they survey are over the limit!
I recall the TGA whining about our fringe labs and the lack of ‘validation’ of our work. I wonder if they will caste the same stones at the FDA?
Before I give this manuscript my usual deep dive and critique, 1st let me congratulate these students for a job well done. I have always joked that DNA quant tools could be run in a High School lab but that was rooted in the fact that after we sold Agencourt, I gave a donation to my High School to ensure their AP bio students would have the equipment to perform DNA sequencing before they graduated. I am passionate about decentralizing genomics as I spent my early career in this field where only the richest most government connected labs could profess to know genomics. Today, USB stick ONT sequencers can be bought for $1K. Qubits are also cheap ($5K) to install in schools.
These students may not have access to all such tools and worked with the BSL-1 White Oak FDA facility.
The students did a great job making very nice figures. I indeed have figure envy and can hopefully use these with citation in the future. There is a joke running in some of my more ornery skeptic circles. Papers with glorious eye candy figures require much deeper dissection:)
The study employed some creative controls. They acquired more research based vaccine preps that were likely to be more contaminated than Pfizer products. They developed a T4 ligation based assay to glue the fragments back into fuller length plasmids to see if any of these could transform E.coli. Some did. Transformation efficiency of E.coli is not the most quantitative tool but it can give you some sense if these are replication competent plasmids in a prep. They found some in the experimental vaccines but none in the approved vaccines.
This may give some assurances that in many vials (the 2 lots tested), there doesn’t appear to be full length plasmids…or least they are not enough of them to transform E.coli after randomly ligating Humpty Dumpty back together again. The heat shock transformation method doesn’t have 100% efficiency and this random ligation assay is a lottery. When you randomly ligate DNA back together, the odds of it assembling back in proper functional order is close to zero. But this was a generous benefit of the doubt experiment.
But we have seen the DNA content vary 1000 fold (10CTs) between lots so this needs to be expanded to the lots that are cooking at CT 13.
The study also used Nanodrops and Agilent Tape stations to triangulate their results. This is good practice and they came to similar conclusions regarding the short comings of each of these tools. I think they are placing too much faith in Agilent to count the number of long molecules.
Nanodrops don’t do a very good job differentiating DNA from RNA or residual nucleotides post DNaseI digestion so take the high end of this paper (470X) with some skepticism. Tape Stations have a hard time quantitating DNA that overlaps with their internal size standards and any long DNA that is a sub-percentage of the total DNA present. Oxford Nanopore (ONT) is the gold standard for length assessments as it can read up to 2Mb (megabases) in length and doesn’t have electrophoresis biases.
The students should know that I have leveled far more criticism at papers published by highly cited PhDs and this is criticism important as this is such a charged topic. Its also a topic many people. even in the biotech field. under appreciate its nuance. Having built some of the more popular DNA purification tools used in the market, I a have become very aware of all of the micro-defects/limitations in various DNA isolation procedures on the market.
Here are my critiques of the paper.
1)The paper does not perform a very balanced review of the literature when it discusses the potential harms of this DNA. No where in the manuscript does it question the 10ng limit (which all vials exceeded) being a limit derived from injected naked DNA. The paper spends a lot of time discussing how small these DNA molecules are and how quickly they ‘should’ be destroyed but doesn’t address the fact that DNA decay curves built on naked DNA cannot be applied to LNP protected DNA. The 10 minute half life of naked DNA in the blood is due to nucleases. These nucleases cannot digest LNP protected DNA so this small size argument is a misdirection. The DNA length required to trigger cGAS-STING is only 20-40bp.
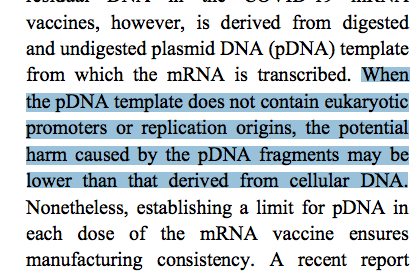
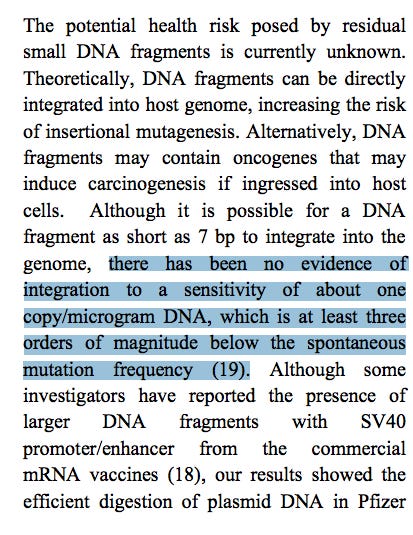
2)The below statement is false. The SV40 Origin of replication and SV40 promoter are active in eukaryotic cells. This should be corrected (not retracted-see conclusions).
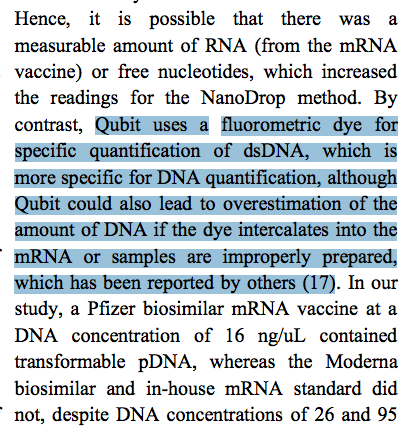
3)The below statement discusses the overestimation that can occur with fluorometry but given they used RNase A, I don’t think RNA signal inflation is what is going on. They should also reference Georgiou et al which demonstrates the picogreen dyes being used in this study will only detect 30% of the DNA when it is treated with DNaseI. To correct for this the 10ng/ul size standards in the Qubit should be DNaseI treated. Do not purify this reaction. Just heat kill the DNaseI or LiDs deactivate it. They could also run a time course on RNAse A digestion to measure if this reaction is complete. It likely is as RNase A is a very active enzyme.
4)The below statement is a misdirection as the study cited (19) is using naked plasmid DNA with an IM injection. These integration frequencies cannot be superimposed onto DNA protected in LNPs that are transfection competent.
Other work published by these same authors from Merck (19) demonstrates these plasmid stick around for 6 months post injection.
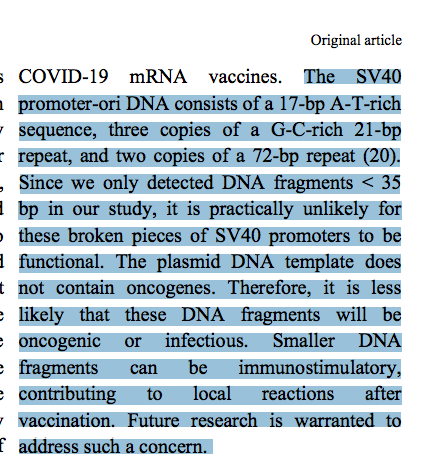
5) Many papers have now found >362bp SV40 fragments (Kammerer et al, Speicher et al). The <35bp measurement should be confirmed with Oxford Nanopore. Our average read lengths on ONT are 214bp with 3.5kb molecules being detected in just 865 reads. The manuscript does not have methods that can reliably comment on fragment size (see below point 6).
This is discussed in this Substack review of the Kammerer paper.
The Wang et al. paper does point out the immuno-stimulatory role of dsDNA transfection. This would be a good time to cite Kwon et al.
I don’t think you can say this DNA doesn’t contain oncogenic DNA.
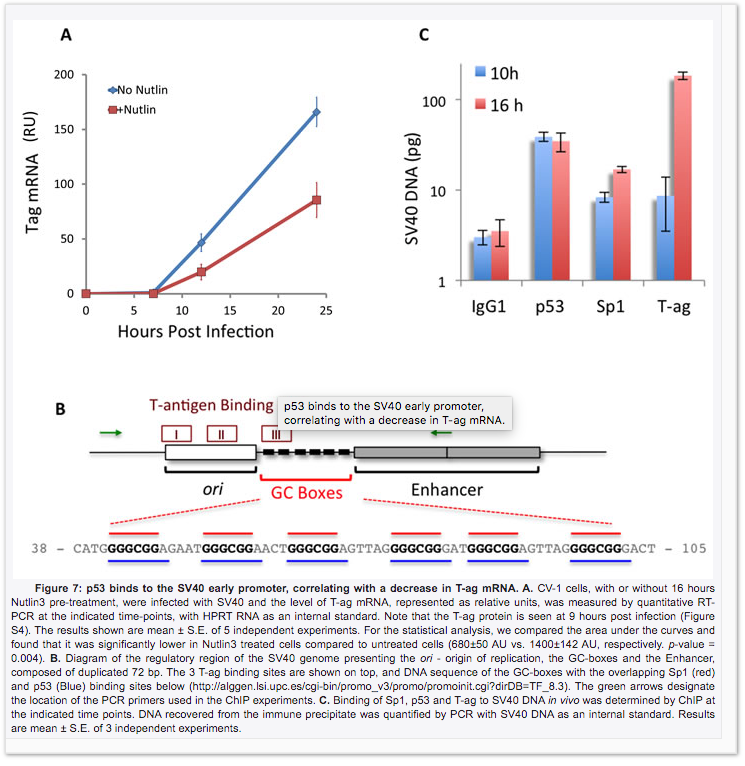
Drayman et al. shows this DNA binds to P53 in figure 7.
Senigl et al. demonstrate SV40 Enhancers are somatic hypermutability elements with potential tumorigenic activity. So the paper shouldn’t be this dismissive of the harms without addressing these papers.
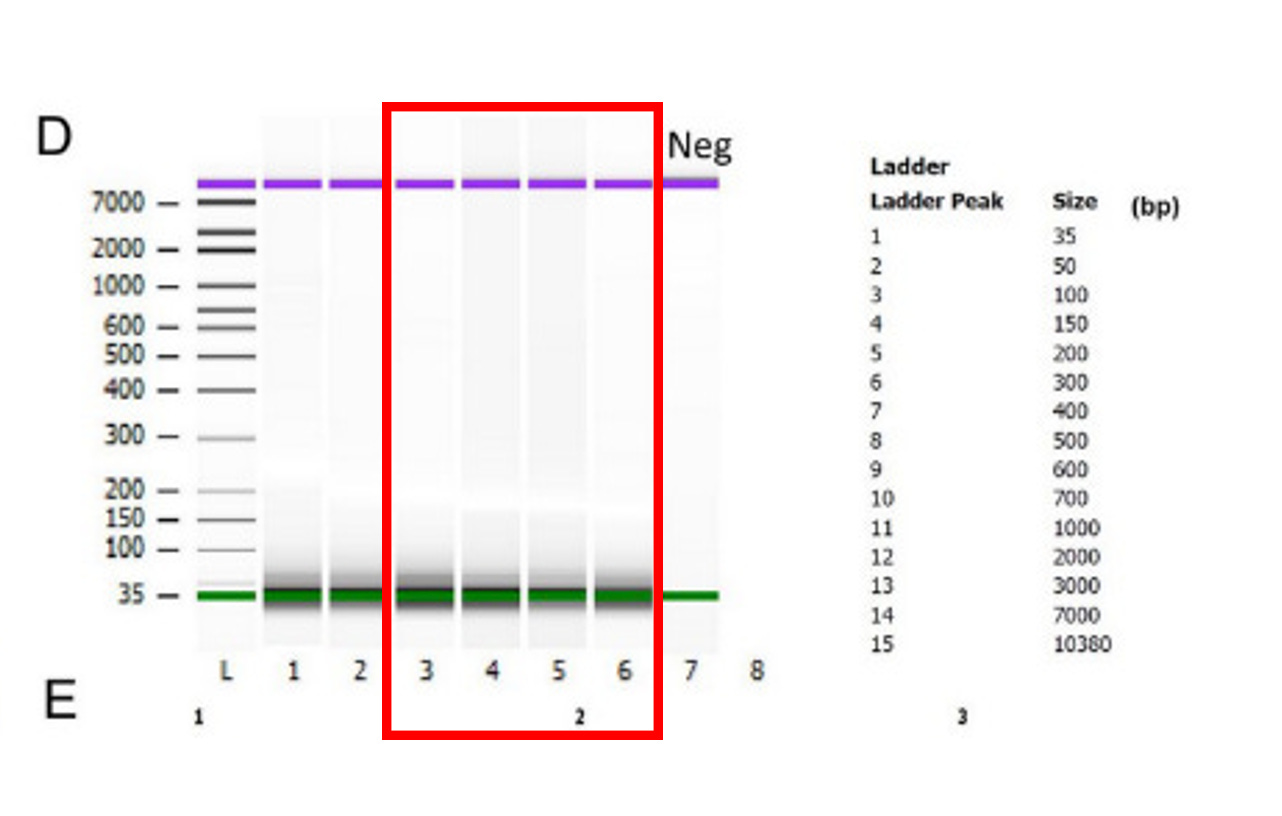
DNA fragment sizing estimates should be confirmed with Oxford Nanopore sequencing. Attempting to size DNA that sits at your lowest size standard on an Agilent Bioanalyzer is error prone. The best method for this is to sequence these molecules so you have single base resolution and confirmation of the sequence of the fragments. When we have done this, 1/400 reads is over 2Kb in size. This is likely hard to visualize on an Agilent Tape station as it doesn’t have very good dynamic range to quantitate molecules at sub percentages of the DNA. They just appear as smears on a gel which can be seen in their data in lanes 4 and 5. Would be nice to have the lot numbers the lanes in Figure D.
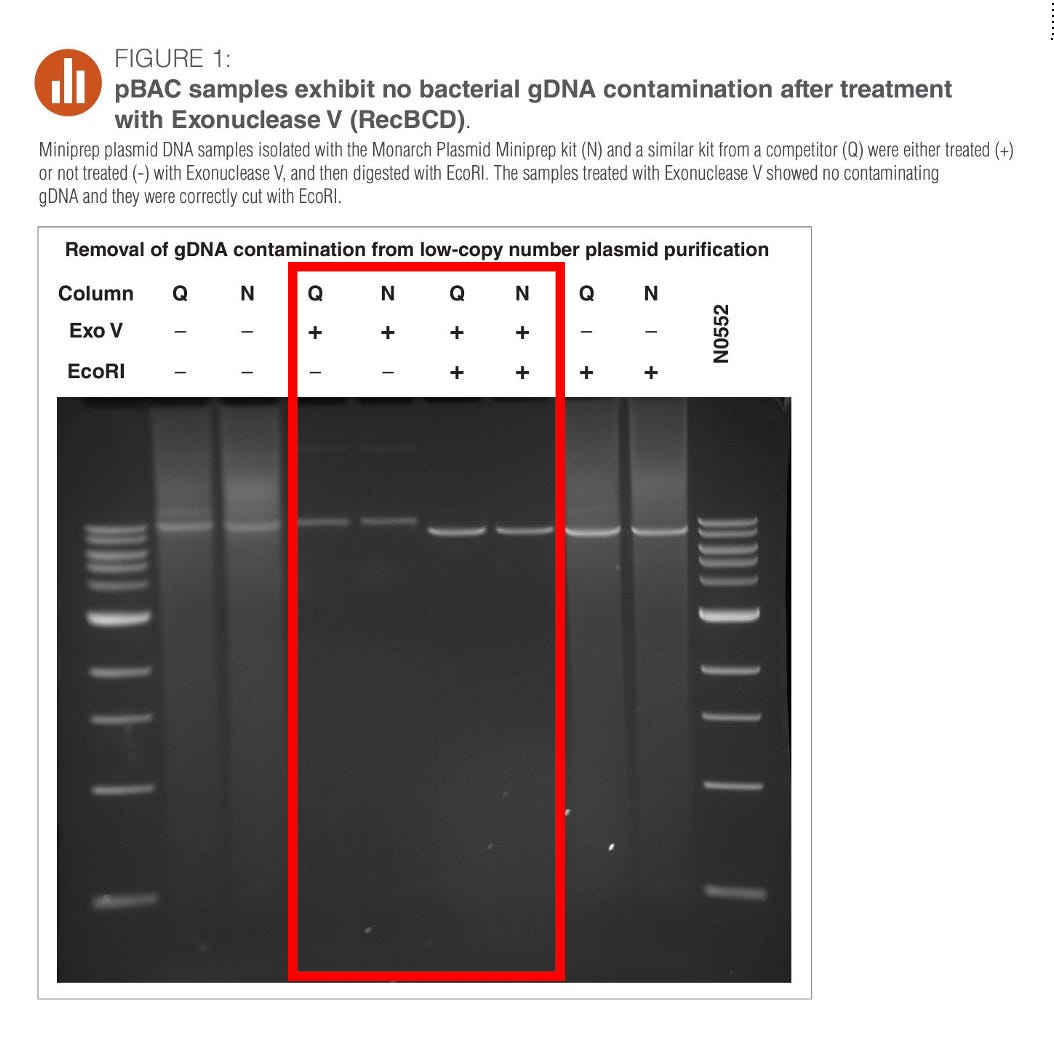
The Methods used to quantitate this DNA has a major flaw which I have described in the below ‘Dirty Deeds’ substack. Wang et al used a NEB Monarch plasmid prep kit which will remove most of the small DNA.
You can see in the below figure from the NEB product guidelines that this plasmid prep system removes the smears (fragmented ssDNA and dsDNA) in the electrophoresis. See the lanes in red with N+. The plasmid is captured but all the small sheared DNA is removed. This significantly impacts the quantitation of small DNA in the vaccines and likely reduces the quantity of the longer and smeared (fragmented) DNA highlighted in red in Figure 5D above.
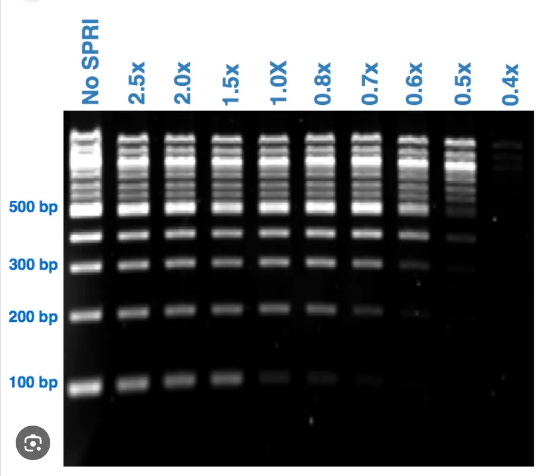
Had the study used a DNA isolation kit that can capture small DNA, their numbers would be even higher! One way to quantitate how much loss they are experiencing with this DNA prep is to run a 10bp DNA ladder through the prep and quantitate how much of each band they capture with the prep compared to the un-prepped ladder.
You can this experiment being performed on SPRI DNA preps with 100bp ladders but for this vaccine application 10bp ladders are in order.
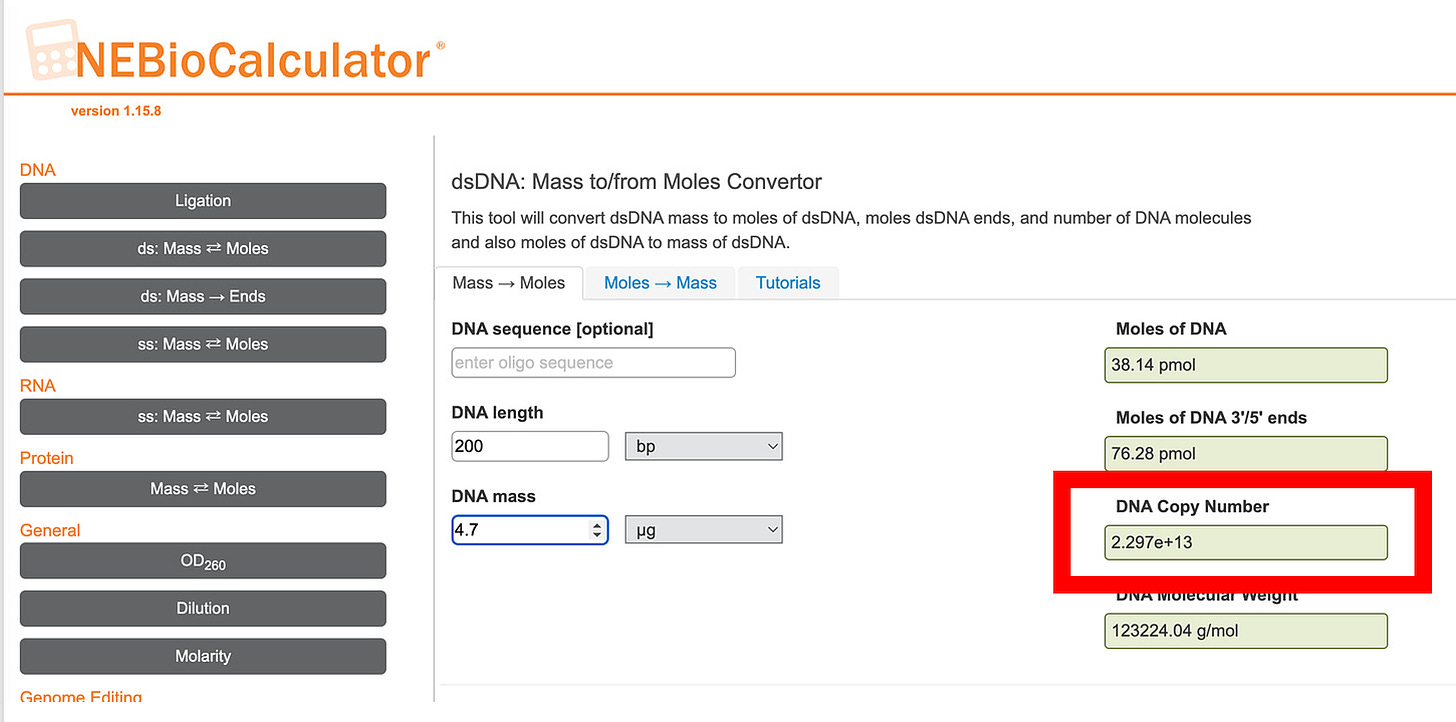
To put this into perspective, lets take the 470X number they cite. This would amount to 4.7ug of DNA. Let’s assume the 200bp fragment size (we found 214bp with ONT) and put that in to NEB’s DNA calculator to count how many DNA molecules we have in the highest point they measured.
23 Trillion fragments of DNA in each dose. If the SV40 Promoter is ~200bp of 7824bp plasmid, that gives us over 500 Billion SV40 promoters per dose. You have 30 trillions cells so each cell could receive 1 plasmid DNA fragment and 1/60th cells could receive one SV40 promoter. In practice, this depends on how many LNPs are in each dose. I’ve seen numbers from 50 Billion-2 Trillion per dose but open to be corrected on this number. The only peer reviewed estimate I have found is in the trillion range per dose.
Keep in mind the higher numbers in this paper are from UV Spec (Nanodrop). The Qubit numbers are closer to 70ng/dose. If we adjust those according to Georgiou et al showing picogreen only picks up 30% of total signal post DNaseI we end up 23X (70ng/0.3 = 233ng/dose) over the limit. Its quite possible the Plasmid Prep also lost 90% of the small DNA which would put this into the 2.0ug/dose (2000ng) range which is more in line with other studies using Qubits without lossy DNA preps (Kammerer, Konig, Speicher, McKernan).
Many will find this number hard to believe as they only get 30ug of RNA per dose and usually T7 polymerase is much more efficient than this. But when you ask T7 Polymerase to incorporate modified nucleotides like N1-methyl-PU, all hell breaks loose.
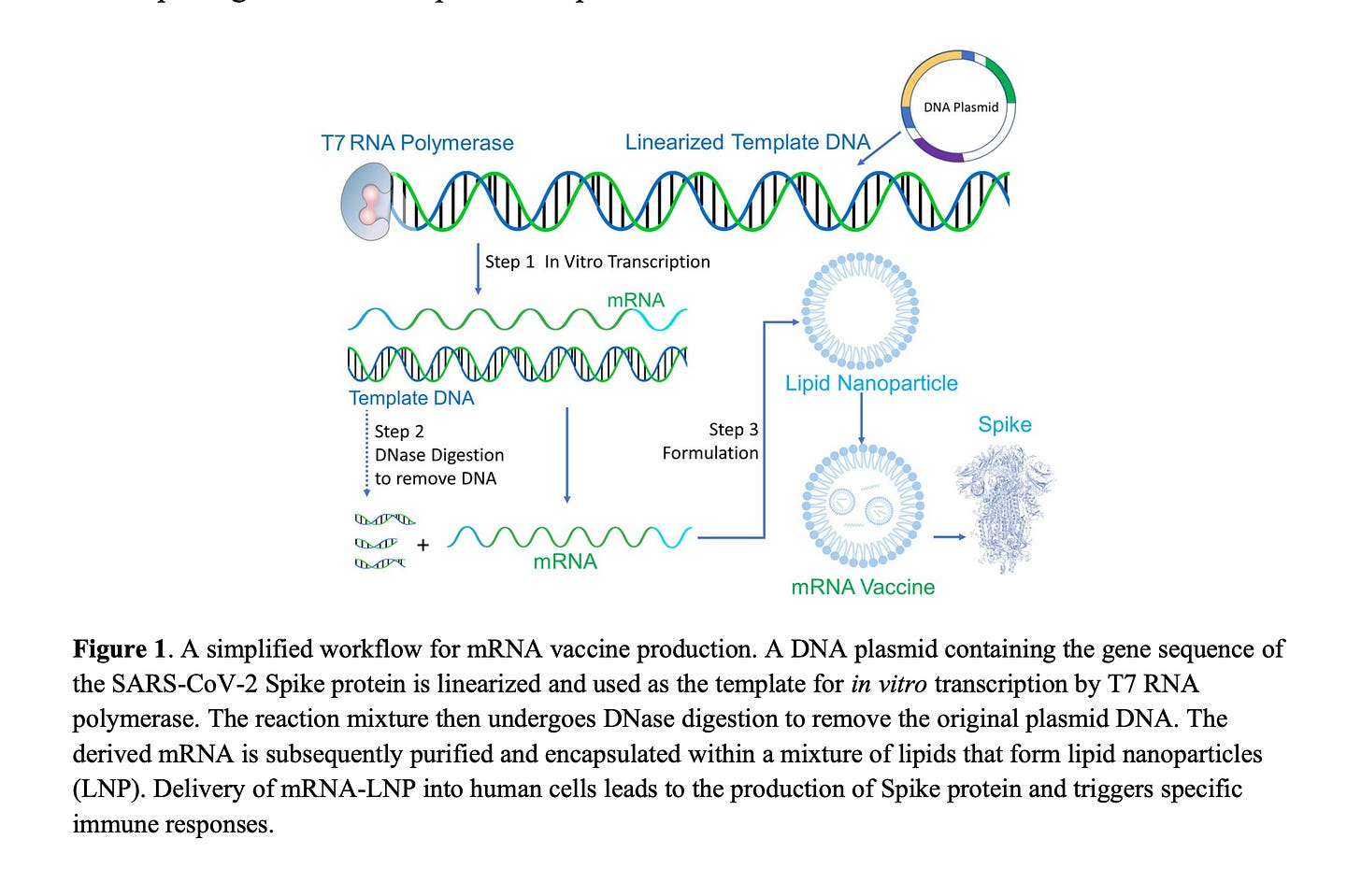
One other technical detail that worth its own deep dive. This very wonderful figure 1 is slightly misleading.
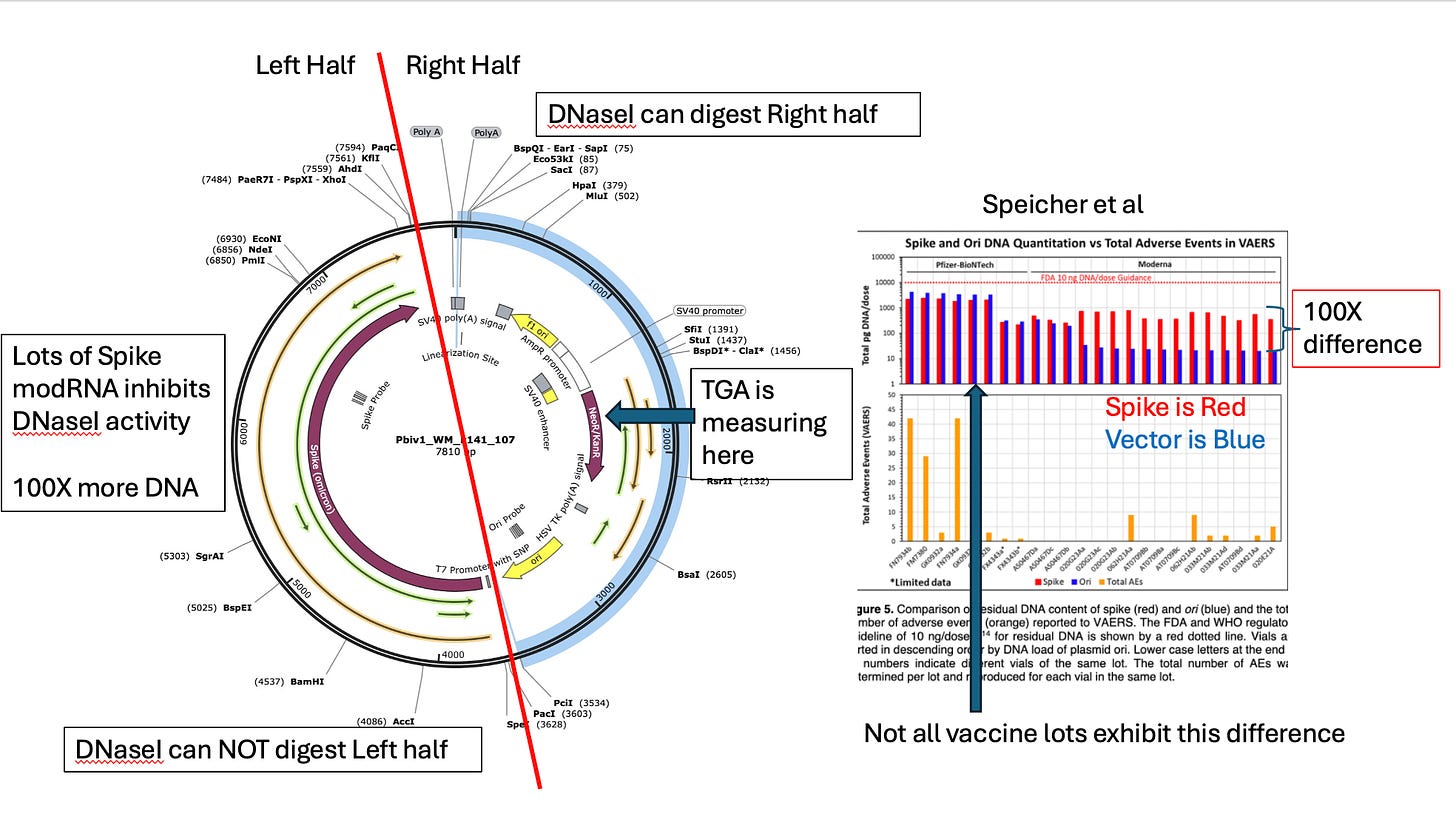
The figure depicts the mRNA and the DNA as being separate on the lower left. The reality is that RNA:DNA hybrids are thermodynamically favored particularly with N1-Methyl-PseudoU. So that modRNA should be decorated with it’s DNA complement. That is why there is so much more DNA in the spike region than in the Kan region. The modRNA is shielding the watson strand of Spike from DNaseI digestion (Sutton et al).
This is also one reason it is foolish to assume the DNA is outside of the LNP while the modRNA is inside of it. They travel together as they are hybridized to each other.
Conclusions
The Wang et al. paper demonstrates even High Schoolers (far above average and impressive) can find this DNA. I don’t know of a science project performed by high schoolers that will be this well cited. Congratulation on a seminal piece of work and ignore the trolls. You will find no shortage of shallow ad hominem attacks and you will have to grow thick skin being on paper that will be read around the world. All papers have limited budgets and shortcomings. Don’t let the above critiques discourage you.
I suspect that some of the softer language in this manuscript that is down playing the significance of the findings, may have had some FDA influence?
Many of these class 1 recalls are for potential risk. I don’t know of anyone that has died from their Hairspray or Costco Eggs. Sure, salmonella is serious but the body count on the mRNA vaccines is off the charts and still no action? The Body count was actually evident in the clinical trials and within 60 days from launch of these vaccines (1223 deaths).
This is in stark contrast to the Swine Flu vaccine that was pulled from the market after just 32 deaths back in the 70s. While many studies recite the ‘Safe and Effective’ mantra, many of these studies have overt conflicts of interest with funding from Pfizer. More independent analysis from Fraiman et al demonstrate 1 Serious Adverse Event for every 800 injections.
When will the FDA break from their Spell? Is this regulatory capture or some ideological re-enactment of the Jonestown massacre?
Let’s not forget that the Data Safety Monitoring Boards for these trials we run by the NIH who now has over $1 Billion in vaccine royalty rolling in. No conflict here.
Good old Anthony Fraudci is rumored to have appointed Richard Whitley from Univ Alabama-Birmingham for this Data Safety Monitoring Board but most other board members remain a secret.
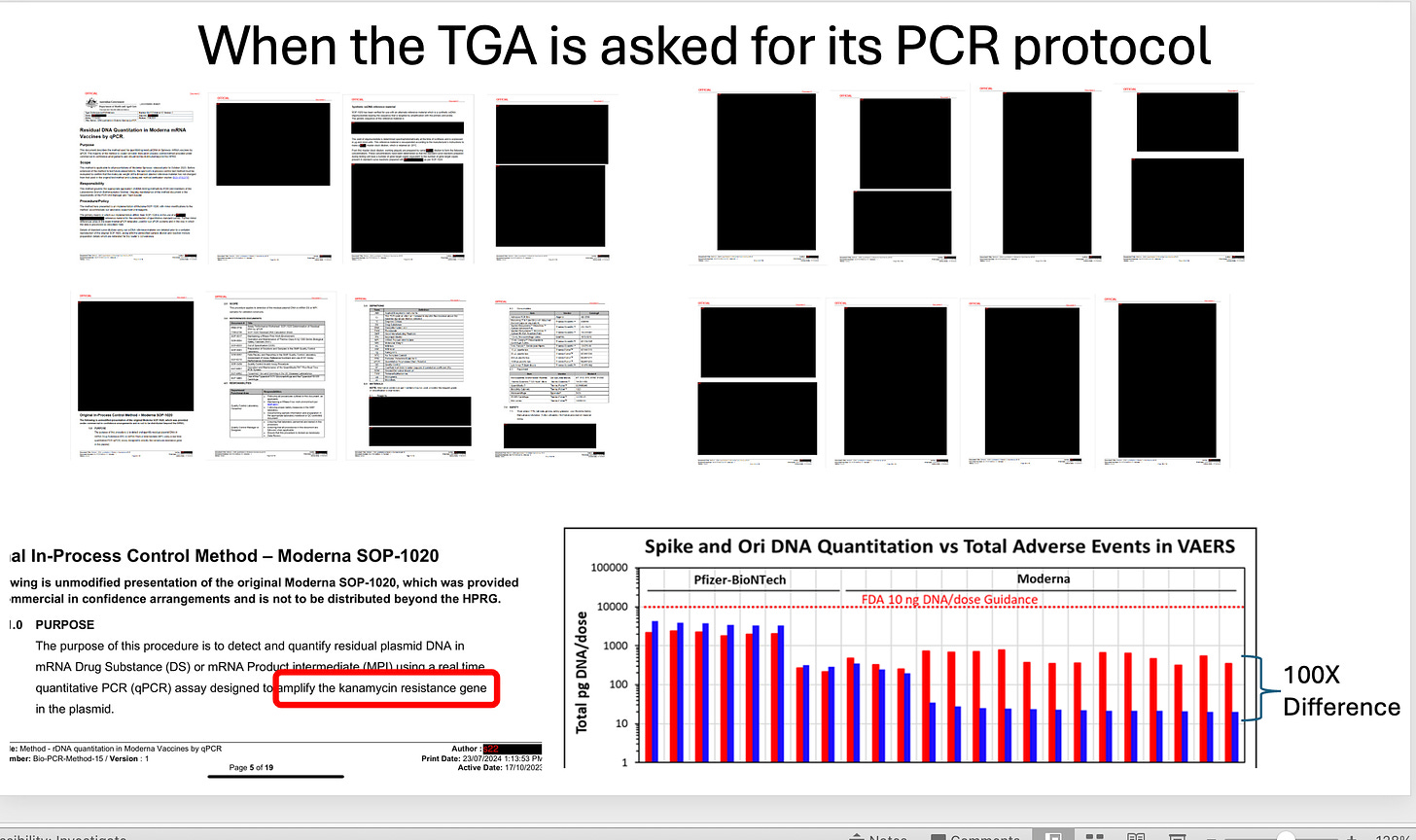
When the TGA is asked for its qPCR protocol used to monitor this problem (now in 3 peer reviewed papers) they return redacted protocols?
One detail they left in the redacted documents is that they are using a single qPCR assay targeting the Kan gene to quantitate the entire plasmid. We know this can give you 100X lower estimate from Speicher’s work.
So there you have it.
Even the FDA’s own staff see the same problem reported by multiple other labs which the Agency at large dismissed. Its important to point out that Keith Pedens (FDA) first knee jerk reaction to our data was that we had a ‘contamination’. Way to pay attention to a whistleblower. Who do you work for? The public or your PDUFA act sponsors?
Now its ~22 months later, and your organization is busy recalling hypothetical Hairspray harms. On June 15th of 2023, I presented this at the VRBAC meeting (4:58:00)
Many more kids were injected with these contaminated products while you were protecting the world from potentially risky hairspray. The real hair on fire moment was given to you 18 months ago.
Maybe all it takes is for the students to become the teacher?
Addendum-
Others have raised calls for the paper to be retracted/reposted due to typos in the lots.
Even with all of the critiques above, I would not encourage retraction as the paper moves the field forward and retraction and reposting will destroy the journal metrics for this work.
I’m generally not a fan of the Nerd Dunking that goes on with the retraction practice. We need to stop viewing papers as binary True or False dichotomies in their entirety.
The solution to error prone speech is more speech, not some label that brands the entire body of work as false because of a typo. Too often pedantic crap like this is weaponized to retract a paper where the errors play no role in the validity or directionality of the paper (Jiang and Mei and Eric Freed).
The criteria for retraction are spelled out in the COPE guidelines. Nothing raised about this paper fits these criteria.
I’m also vested in teaching High School students genomics and I find some academics take sadistic pleasure trolling the new entrants to the field as if they are some Fraternity pledge slave. PhD/PostDoc culture very much does this to grown adults. A retraction can be a career ending move. To suggest this on High School authors for typos before suggesting an erratum is cold as hell nerd dunking. Lets teach the next generation and not play the cancel culture/censorship games we witness with many of the vaxophiles during the pandemic.
A second critique of the work is grounded in a False Dichotomy. “DNA contamination work comes at the cost of focusing on other more serious risks”. It is possible to walk and chew gum at the same time. I have been very clear in most of my presentations on this that removal of the DNA from these shots will not magically make them safe as I don’t think you can address rapidly mutating RNA viruses with vaccine programs any more than you should supply everyone with antibiotics at the same time. Bacteria mutate slower than RNA viruses so the risk of resistance is much more real with narrow Vaccines that attempt to suppress a RNA virus. Mucosal immunity through injection is also specious.
This false dichotomy is very different than the dichotomy that unfolded on Twitter this week with Calley Means in the famous Fruit Loops vs SV40 debate on the Danny Jones Podcast with Dr. Mary Talley Bowden and Dr. Jack Kruse. In this scenario, Calley wont mention vaccines in his book nor call for the removal of the shots from the market (despite not giving them to his kids) which presents an Either/OR logic into the discussion when AND or ‘both’ is the more rational approach. Even if you pursue both, your priority should be on the products causing imminent death.