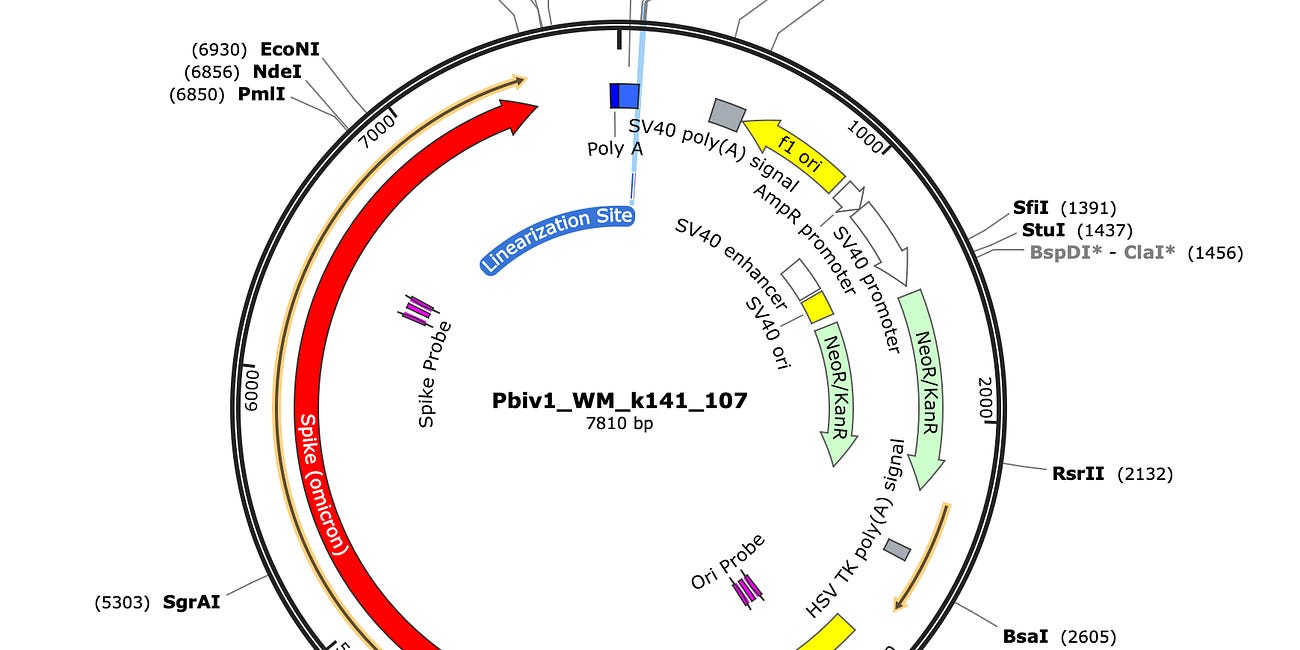
SV40 origin of replication in mammalian cells in absence of SV40 Large T-Antigen
Positive tumor biopsy qPCR one year after vaccination
We’ve previously described BNT162b2 transfected OVCAR3 cell lines showing evidence of BNT162b2 plasmid DNA replication. This is perplexing as SV40 origins of replication usually require SV40 Large Tumor-Antigen to replicate in mammalian cells and Pfizer vaccines do not contain SV40 Large T-Antigen.
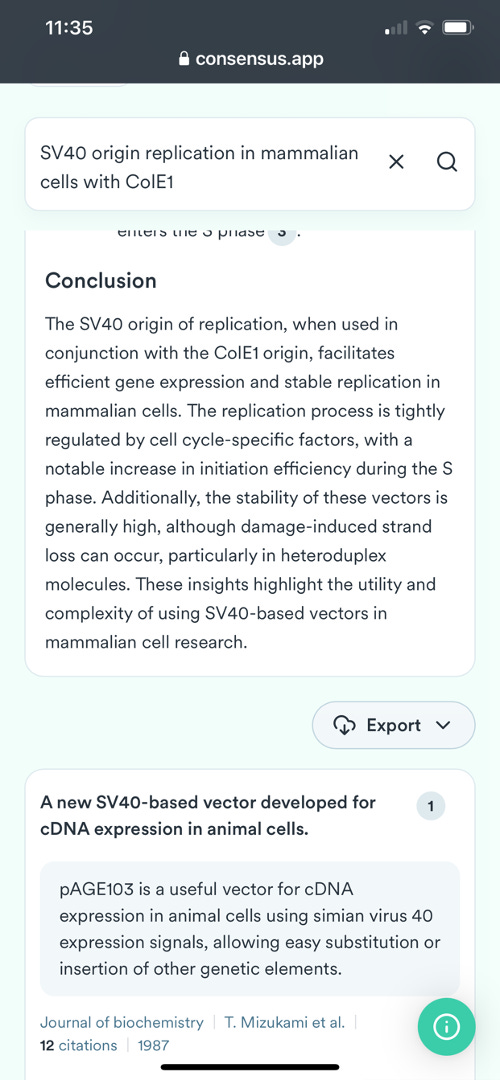
This led me to test out a new LLM tool known as Consensus.app which is like a ChatGPT tuned for literature searching. Nick Jikomes turned me onto this-
This LLM quickly delivered the answer.
Links to a few references it cites.
For those not familiar with the Pfizer vaccine it contains 3 origins of replication: a SV40 ori, a ColE1 ori and a F1 origin of replication. Consensus.app found a few papers that demonstrate SV40 Origins of replication can replicate in mammalian cells without SV40 Large Tumor Antigen. The requirements are a ColE1 and F1 origin, both of which exist in the Pfizer plasmid. A 3rd mechanism of SV40 ori replication requires S/MARs domains which are found in Beta Globin sequences. The Pfizer vaccine also has beta globin UTRs which need further exploring regarding their capacity to emulate a S/MAR domain.
Now that we have a mechanism for replication, we can better explain the evidence we have collected. For instance, this would explain the variants we are seeing in the vaccine only when it is transfected into human cell lines. When we sequence the naked vaccine, we see no variants in the plasmid backbone. When we sequence the vaccine after it has been transfected into OVCAR3 cells, we see SNPs in the origins of replication.
This has been perplexing as we would not expect to see such high frequency variants (above 10% of the reads showing variants at a given SNP) from polymerase replication errors alone. Polymerase replication errors are usually 1 in a 10,000 to a 1 in a million in frequency and we don’t have the depth of sequencing to see those. Some other mechanism must be making these high prevalence (14/100 reads confirming the SNP). These Consensus.app delivered papers reveal a mechanism.
Its important to steel man this.
Some have suggested the pUC ori’s (ColE1) used to express the polymerases in our PCR kits could be contributing to the detection of some SNPs under the ColE1 ori. This is plausible but our kits usually have NTC signals out past 35 CTs for the ColE1 ori primer set. We are only sequencing samples that have CTs <28 (usually less than 22). We wouldn’t expect to see >10% allele frequency in IGV with a 35CT contaminant (>7deltaCT is 1/128th the load) . We also see these SNPs in the SV40 origin and the F1 origin which don’t suffer from being in the background of polymerase kits.
This suggests the errors are from replication and these papers imply the regions most prone to replication errors are in heteroduplexes often found in Origins of replication. This best fits the data we have observed to date.
In summary
1)The vaccine is clearly making it into the cells or these mutations would not be seen in our sequencing data. When we sequence the naked vaccine alone as a control we see no variants. Once that vaccine is transfected into human cells variants emerge.
2)The DNA is replicating and until I played around with Consensus.app I didn’t have a good explanation as to why this could occur in absence of SV40 Large T-Antigen.
3)If the DNA is replicating in mammalian cells, then we don’t need self amplifying mRNA vaccines as the population was already given them with Pfizer vaccines.
What are the implications of injected DNA that can copy itself?
First and foremost, the FDA regulations speaking to 10ng of DNA are now irrelevant as you can drive a truck through that regulation with LNPs containing DNA that has active mammalian origins of replication.
I was always skeptical of the shedding literature. Not because we lack anecdotes or good explanations of the hypothesis as put forth by Dr. Pierre Kory and others,
but because I just couldn’t wrap my head around the dosage. Even if we do shed small amounts of spike coated exosomes as suggested by Bansal et al, the dose of Spike to the recipient would be so minuscule, how could it create disease in a recipient? I also think much of the damage from vaccination (but not all) is related to the Bolus theory put forward by
. Shedding recipients are not exposed to this risk factor.The dosage calculus completely changes if any of these exosomes contain self replicating DNA or plasmids. Small amounts of shedded plasmids could expand in the recipient and perhaps explain this phenomenon.
If the DNA is replicating in cells, it has profound implications regarding the DNA persistence in vaccinated patients. Here is an case where a lab plasmid created a casedemic triggering FP qPCR tests for nucleocapsid. The Pfizer plasmids are designed to also replicate in E.coli like this paper.
Curious case of nucleocapsid encoding Plasmids colonizing Lab staff
How long does this replication competent vaccine DNA last in patients?
We are in the process of running qPCR for vaccine DNA in vaccinated patients’ tumor samples. We are finding qPCR positive samples 1 year after vaccination that are equal to or in more quantity than the RNAP human genomic control.
What does that mean? RNAP is common qPCR assay that targets a human gene. It is used in some COVID 19 PCR kits as an internal control. It was notably absent in the sloppy Corman-Drosten PCR test making that test clinically negligent as you had no way to compare the viral RNA to the number of human cells collected with a nasal swab. Since nasal swabs can vary 1000 fold in the number of human cells collected, no viral load could be calculated without an RNAP control.
If you qPCR for SV40 from the Pfizer vaccine and it is at the same or higher copy number than the human RNAP gene in the sample a year later, then the SV40 DNA has not been diluted out over time and appears to be replicating with the genomic DNA. This is a sign of either integration or episomal replication of the SV40 DNA with each cell division.
This is not an expected result. Lets go over some simple dilution math on the volumes involved here.
This patient took 4 Pfizer vaccines. His last one was 1 year before these biopsies were taken. He died ~30 days after this biopsy.
Given 300ul-1200ul (1-4 Pfizer doses) of vaccine are diluted into 87,000 ml of body volume (Average human body volume is 87 Liters), we should see a dilution of vaccine DNA of at least 64,000 fold on day 1 if the vaccine is evenly distributed to the entire body volume. If the DNA stayed in the arm as advertised, we should see no DNA in Colon tumor biopsies.
A 64,000 fold dilution would manifest in a tissue that is 16 CT later (2^16) signal in qPCR compared to the original DNA signal found in the 300ul vaccine.
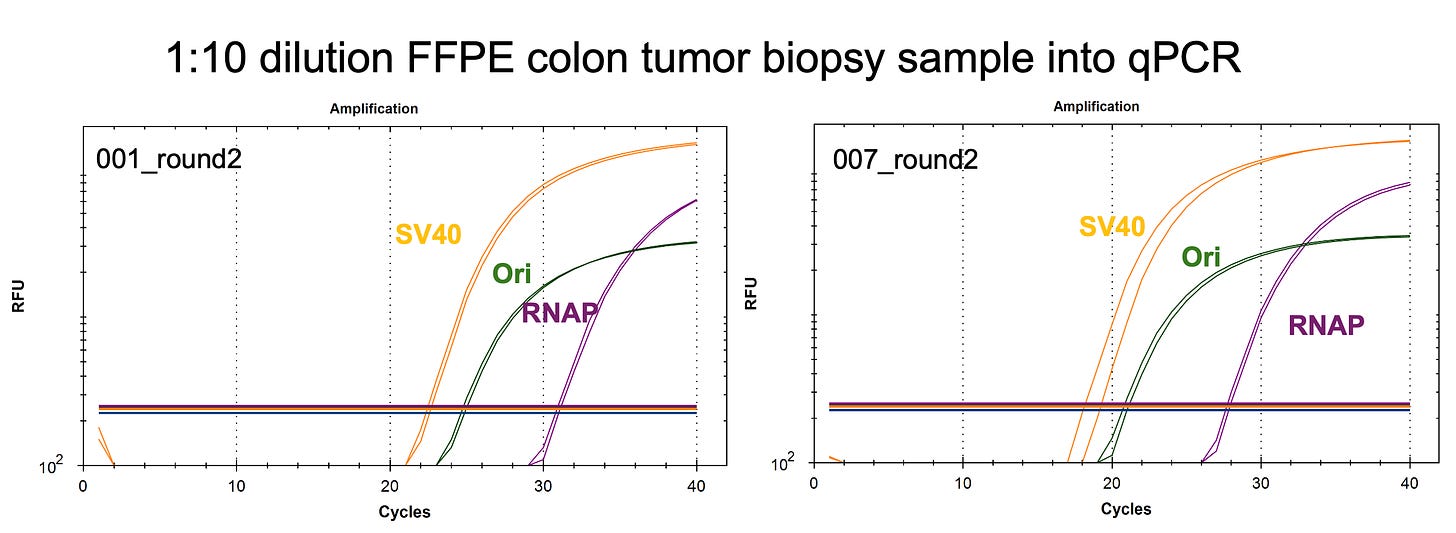
Yet we are finding tissues that have similar CTs to the naked vaccine a year later. That can only happen if the vaccine DNA is integrating and amplifying or if the DNA is being replicated by these origins of replication as episomal plasmids.
If it were vaccine contamination from our lab or the lab collecting the specimens (INMODIA GmbH), we should see equal CTs for Spike, SV40 promoter and ColE1 Ori as the vaccine contains equal quantities of all 3 targets in most Pfizer vaccines. The spike assay, surprisingly fails to detect any DNA in this biopsy which is unusual as spike DNA is sometimes 6CTs (earlier) higher in concentration in some Moderna lots due to incomplete DNaseI treatment of RNA:DNA hybrids. Speicher et al demonstrate this artifact but it is not present in Pfizers manufacturing process in the vials we have surveyed to date.
Instead, we observe SV40 promoter and ColE1 Ori offsets with no spike signal and the NTCs are clean out to CT 35.
Lab vaccine contamination would also not deliver the variants we are seeing in the sequence data. As demonstrated in our OVCAR3 study, If you simply mix naked vaccine DNA with FFPE gDNA and sequence them, you don’t see the SNPs the cells are making. We have a very specific signature that this DNA has been modified by transfection into cells in a way I don’t think any researcher could hack together to create a scam.
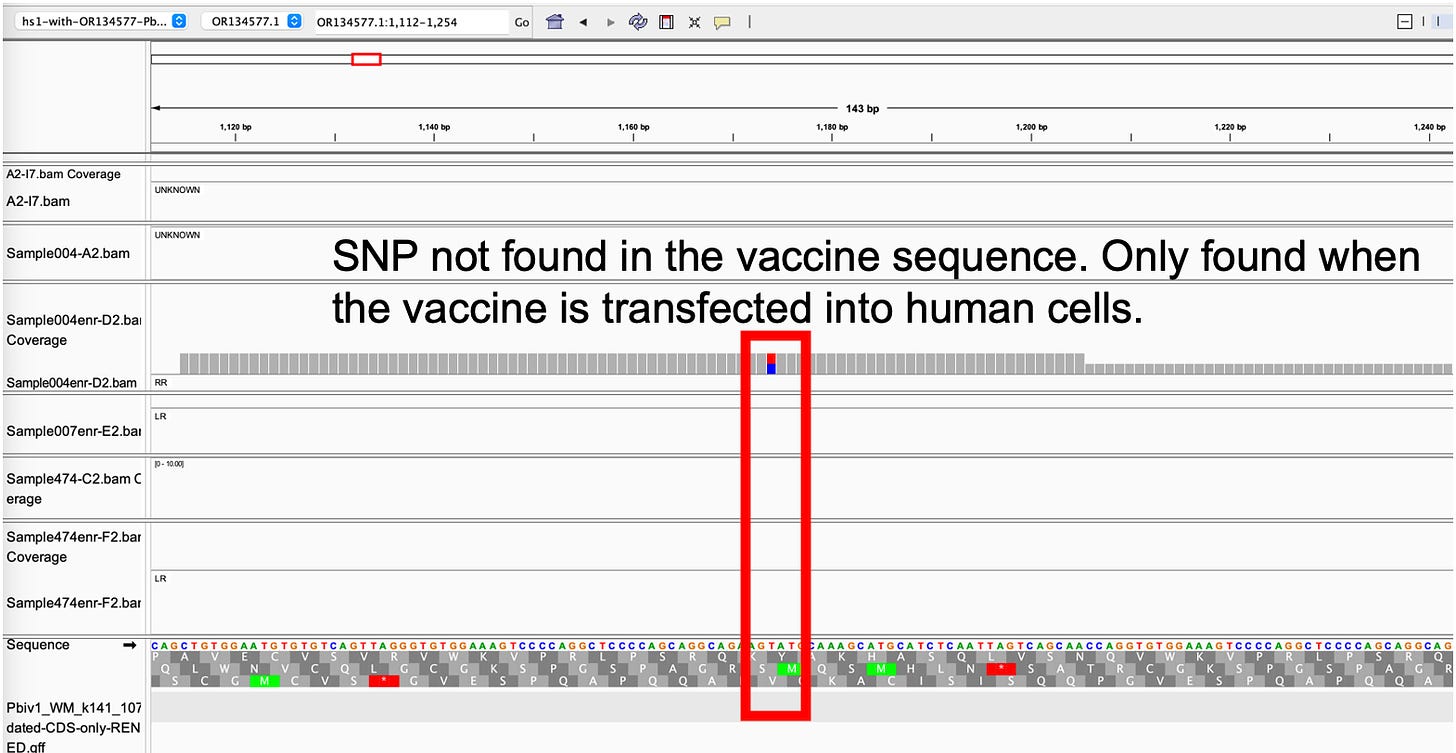
Why are we currently seeing only SV40 promoter and ColE1 Ori qPCR?
It is possible some vials contain small portions of empty vector. This is contaminating DNA where the spike insert has failed to be cloned into the plasmid. It is also possible that portions of the spike sequence have been deleted through plasmid replication with ColE1 origins of replication that are notorious for rearrangements with larger insert plasmids. We do see some spike sequence in the data but we don’t have the depth of coverage to ascertain what has occurred in the region of the spike sequence that our qPCR assay targets.
TL/DR
Pfizer vaccine sequence can be detected 1 year after vaccination in a spike positive IHC colon cancer biopsy. The longest detection of nucleic acids post vaccination is via Roltgen (60days).
The very low CTs for these samples can only be explained by replication competency of the vaccine DNA or genome integration and amplification. Preliminary sequencing of these samples confirms the qPCR is not off target signal and that variants exist in the plasmids. This is indicative of replication errors or biotransformation of the vaccine DNA. Deeper sequencing is being performed to resolve integration or episomal replication of the vaccine DNA.
Some details of the sequencing experiment
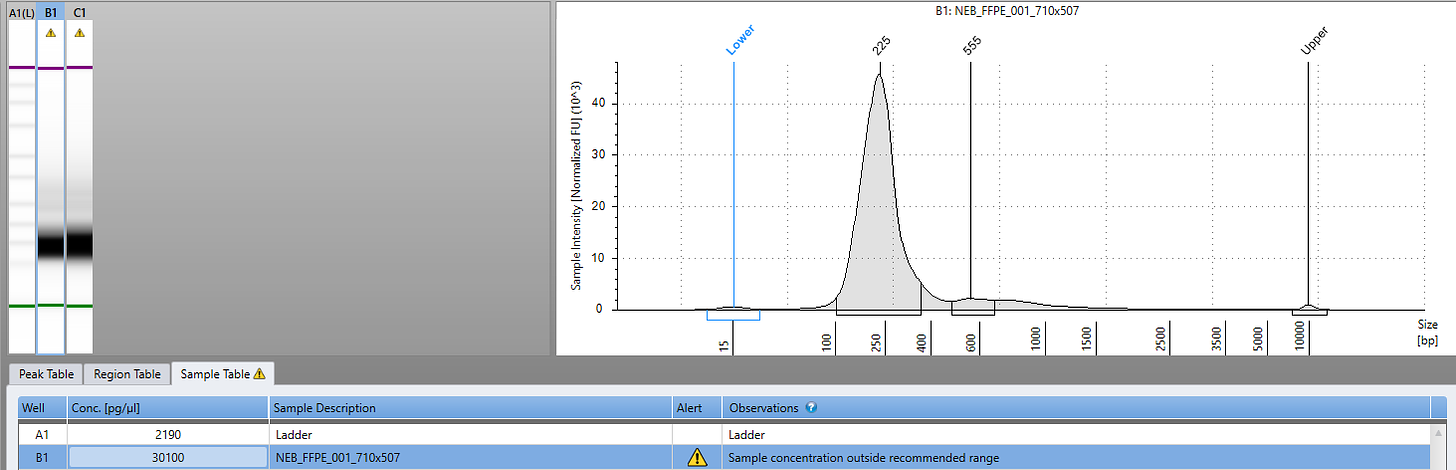
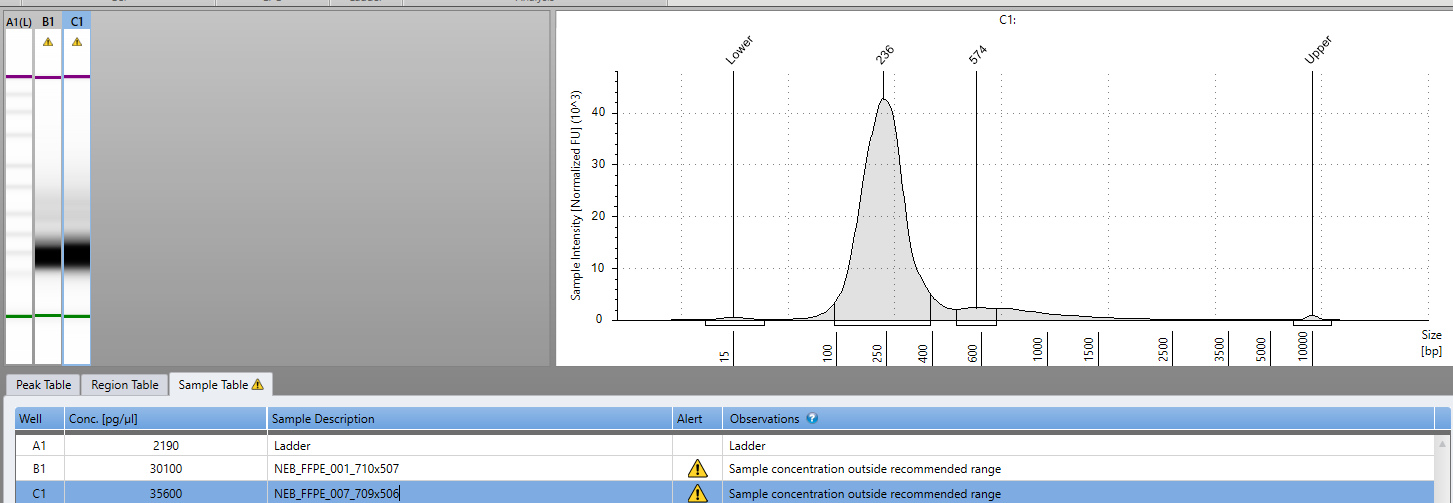
We performed 0.6X sequence coverage of this sample to assess how well this FFPE DNA was going to sequence. FFPE DNA is notorious to sequence as its often cross linked and contains abasic sites.
The sequencing had high levels of small inserts (fragmented DNA) unlikely to find integration events. We have since constructed new sequencing libraries with NEBs latest FFPE library construction kit. This kit repairs abasic sites in the DNA before constructing Illumina sequencing libraries and with just 8 cycles of PCR we have 30ng/ul in 30ul of library for biopsies taken before and after death of the patient.
A portion (20ul) of DNA from both of these libraries was pooled and used with IDT xGEN lockdown probe enrichment method. This process ‘fish hooks’ out all of the DNA that shares sequence with the Pfizer and Moderna plasmids so the sequencer can be focused on plasmid like sequences. It is meant to provide higher depth of sequencing coverage over the regions of the sample that share sequence homology with the vaccine.
Briefly, 214 biotinylated 100mer probes were synthesized and used as hybridization capture reagents. Once hybridized to your Illumina library, streptavidin magnetic beads bind the biotin and a magnet is used to harvest the vaccine-like sequences.
Methods
Colon tumor biopsies that were spike IHC positive were selected for DNA analysis. DNA was purified by INMODIA, GmbH and sent to Medicinal Genomics in Beverly Mass for qPCR and sequence analysis.
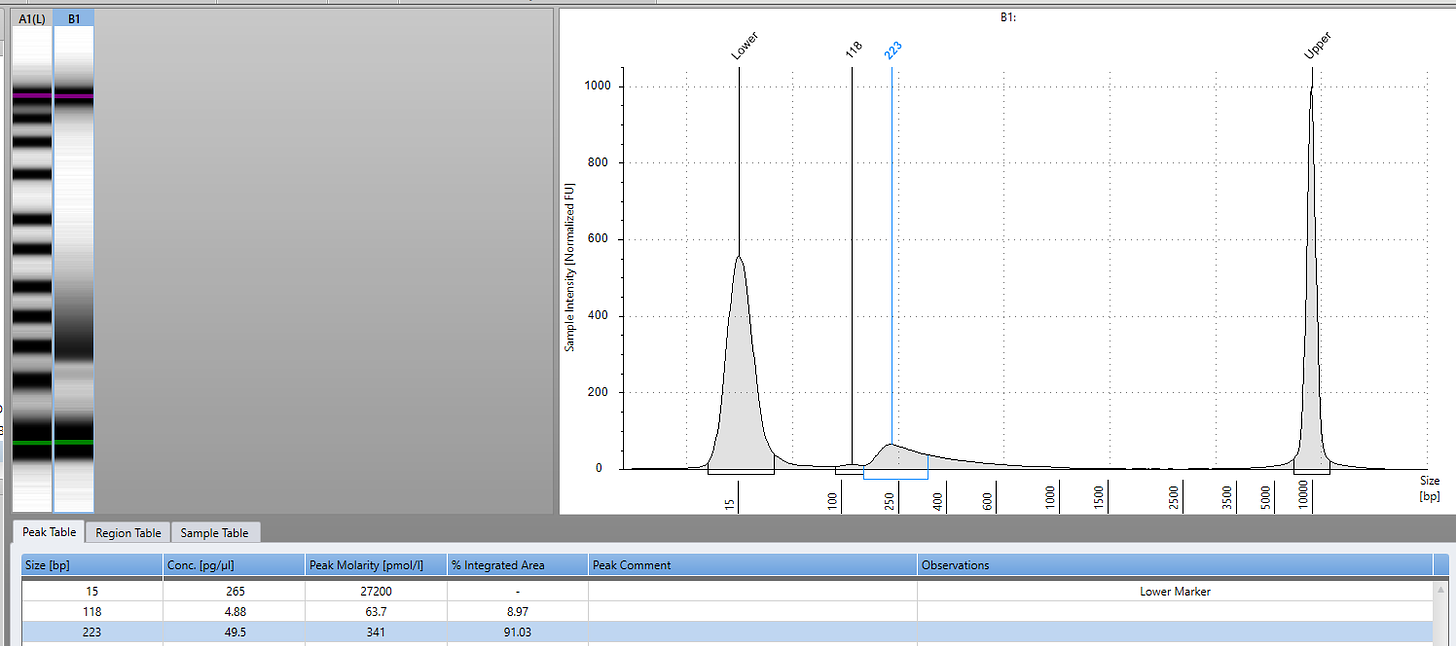
2ul of 1:10 diluted DNA was used in a 18ul qPCR assay previously described. Positive samples were prioritized for sequencing using NEBNext UltraShear™ FFPE DNA Library Prep Kit (NEB #E6655). 14ul of FFPE DNA was diluted into 15ul ddH20 for 29ul of starting input to the library preparation process. 8 cycles of PCR was utilized to amplify the final library with NEBNext® Multiplex Oligos for Illumina® (#E7600S).
PCR samples were purified using 0.9X Ampure and evaluated on an Agilent Tape Station for insert size and quantity.
Sequencing was performed on as 2x150bp reads on an Illumina NovaSeq with dual index barcodes.
Analysis
Reads were trimmed with Trimmomatic and mapped to GRCh38.p14 using DRAGEN mapper from Illumina. Read mappings were visualized in IGV.






“ If the DNA is replicating in mammalian cells, then we don’t need self amplifying mRNA vaccines as the population was already given them with Pfizer vaccines.” omg
the technical content is way above my paygrade but the conclusions are not...
remains the question : was it an accident or was it on purpose???